Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance
- PMID: 27595477
- PMCID: PMC5957269
- DOI: 10.1038/ng.3659
Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance
Erratum in
-
Corrigendum: Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance.Nat Genet. 2017 Jan 31;49(2):317. doi: 10.1038/ng0217-317a. Nat Genet. 2017. PMID: 28138154 No abstract available.
Abstract
Esophageal adenocarcinoma (EAC) has a poor outcome, and targeted therapy trials have thus far been disappointing owing to a lack of robust stratification methods. Whole-genome sequencing (WGS) analysis of 129 cases demonstrated that this is a heterogeneous cancer dominated by copy number alterations with frequent large-scale rearrangements. Co-amplification of receptor tyrosine kinases (RTKs) and/or downstream mitogenic activation is almost ubiquitous; thus tailored combination RTK inhibitor (RTKi) therapy might be required, as we demonstrate in vitro. However, mutational signatures showed three distinct molecular subtypes with potential therapeutic relevance, which we verified in an independent cohort (n = 87): (i) enrichment for BRCA signature with prevalent defects in the homologous recombination pathway; (ii) dominant T>G mutational pattern associated with a high mutational load and neoantigen burden; and (iii) C>A/T mutational pattern with evidence of an aging imprint. These subtypes could be ascertained using a clinically applicable sequencing strategy (low coverage) as a basis for therapy selection.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
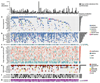




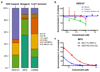

Comment in
-
Oesophageal cancer: Defining tumour subtypes in oesophageal adenocarcinoma.Nat Rev Gastroenterol Hepatol. 2016 Oct;13(10):557. doi: 10.1038/nrgastro.2016.157. Epub 2016 Sep 14. Nat Rev Gastroenterol Hepatol. 2016. PMID: 27625190 No abstract available.
References
-
- Ferlay J, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International Journal of Cancer. 2015;136(5):E359–86. - PubMed
-
- Cunningham D, Okines AF, Ashley S. Capecitabine and oxaliplatin for advanced esophagogastric cancer. N Engl J Med. 2010;362(9):858–9. - PubMed
-
- Allum WH, et al. Long-term results of a randomized trial of surgery with or without preoperative chemotherapy in esophageal cancer. J Clin Oncol. 2009;27(30):5062–7. - PubMed
-
- Cunningham D, et al. Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. N Engl J Med. 2006;355(1):11–20. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

