Viruses-to-mobile genetic elements skew in the deep Atlantis II brine pool sediments
- PMID: 27596223
- PMCID: PMC5011723
- DOI: 10.1038/srep32704
Viruses-to-mobile genetic elements skew in the deep Atlantis II brine pool sediments
Abstract
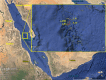
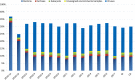
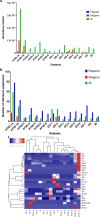
The central rift of the Red Sea has 25 brine pools with different physical and geochemical characteristics. Atlantis II (ATIID), Discovery Deeps (DD) and Chain Deep (CD) are characterized by high salinity, temperature and metal content. Several studies reported microbial communities in these brine pools, but few studies addressed the brine pool sediments. Therefore, sediment cores were collected from ATIID, DD, CD brine pools and an adjacent brine-influenced site. Sixteen different lithologic sediment sections were subjected to shotgun DNA pyrosequencing to generate 1.47 billion base pairs (1.47 × 10(9) bp). We generated sediment-specific reads and attempted to annotate all reads. We report the phylogenetic and biochemical uniqueness of the deepest ATIID sulfur-rich brine pool sediments. In contrary to all other sediment sections, bacteria dominate the deepest ATIID sulfur-rich brine pool sediments. This decrease in virus-to-bacteria ratio in selected sections and depth coincided with an overrepresentation of mobile genetic elements. Skewing in the composition of viruses-to-mobile genetic elements may uniquely contribute to the distinct microbial consortium in sediments in proximity to hydrothermally active vents of the Red Sea and possibly in their surroundings, through differential horizontal gene transfer.
Figures

Similar articles
-
Insertion sequences enrichment in extreme Red sea brine pool vent.Extremophiles. 2017 Mar;21(2):271-282. doi: 10.1007/s00792-016-0900-4. Epub 2016 Dec 3. Extremophiles. 2017. PMID: 27915389
-
Unique prokaryotic consortia in geochemically distinct sediments from Red Sea Atlantis II and discovery deep brine pools.PLoS One. 2012;7(8):e42872. doi: 10.1371/journal.pone.0042872. Epub 2012 Aug 20. PLoS One. 2012. PMID: 22916172 Free PMC article.
-
Hydrothermally generated aromatic compounds are consumed by bacteria colonizing in Atlantis II Deep of the Red Sea.ISME J. 2011 Oct;5(10):1652-9. doi: 10.1038/ismej.2011.42. Epub 2011 Apr 28. ISME J. 2011. PMID: 21525946 Free PMC article.
-
An abyssal mobilome: viruses, plasmids and vesicles from deep-sea hydrothermal vents.Res Microbiol. 2015 Dec;166(10):742-52. doi: 10.1016/j.resmic.2015.04.001. Epub 2015 Apr 22. Res Microbiol. 2015. PMID: 25911507 Review.
-
Horizontal gene transfer and mobile genetic elements in marine systems.Methods Mol Biol. 2009;532:435-53. doi: 10.1007/978-1-60327-853-9_25. Methods Mol Biol. 2009. PMID: 19271200 Review.
Cited by
-
Insertion sequences enrichment in extreme Red sea brine pool vent.Extremophiles. 2017 Mar;21(2):271-282. doi: 10.1007/s00792-016-0900-4. Epub 2016 Dec 3. Extremophiles. 2017. PMID: 27915389
-
Antibiotic resistance in the most unlikeliest of places.Microb Biotechnol. 2017 Nov;10(6):1454-1456. doi: 10.1111/1751-7915.12868. Epub 2017 Oct 13. Microb Biotechnol. 2017. PMID: 29027382 Free PMC article. No abstract available.
-
Insights into Red Sea Brine Pool Specialized Metabolism Gene Clusters Encoding Potential Metabolites for Biotechnological Applications and Extremophile Survival.Mar Drugs. 2019 May 8;17(5):273. doi: 10.3390/md17050273. Mar Drugs. 2019. PMID: 31071993 Free PMC article.
-
The association of group IIB intron with integrons in hypersaline environments.Mob DNA. 2021 Mar 1;12(1):8. doi: 10.1186/s13100-021-00234-2. Mob DNA. 2021. PMID: 33648565 Free PMC article.
-
Active prokaryotic and eukaryotic viral ecology across spatial scale in a deep-sea brine pool.ISME Commun. 2024 Jun 14;4(1):ycae084. doi: 10.1093/ismeco/ycae084. eCollection 2024 Jan. ISME Commun. 2024. PMID: 39021441 Free PMC article.
References
-
- Blanc G., Anschutz P. & Pierret M. C. In Sedimentation and Tectonics in Rift Basins Red Sea:- Gulf of Aden (eds BruceH Purser & Bosence DanW J.) Ch. 27, 505–520 (Springer: Netherlands, 1998).
-
- Gurvich E. G. In Metalliferous Sediments of the World Ocean Ch. 3, 127–210 (Springer: Berlin Heidelberg, 2006).
-
- Swallow J. C. & Crease J. Hot Salty Water at the Bottom of the Red Sea. Nature 205, 165–166 (1965).
-
- Bosworth W., Huchon P. & McClay K. The Red Sea and Gulf of Aden Basins. Journal of African Earth Sciences 43, 334–378, 10.1016/j.jafrearsci.2005.07.020 (2005). - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

