G-quadruplex prediction in E. coli genome reveals a conserved putative G-quadruplex-Hairpin-Duplex switch
- PMID: 27596596
- PMCID: PMC5100583
- DOI: 10.1093/nar/gkw769
G-quadruplex prediction in E. coli genome reveals a conserved putative G-quadruplex-Hairpin-Duplex switch
Abstract
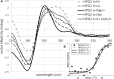
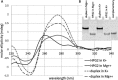
Many studies show that short non-coding sequences are widely conserved among regulatory elements. More and more conserved sequences are being discovered since the development of next generation sequencing technology. A common approach to identify conserved sequences with regulatory roles relies on topological changes such as hairpin formation at the DNA or RNA level. G-quadruplexes, non-canonical nucleic acid topologies with little established biological roles, are increasingly considered for conserved regulatory element discovery. Since the tertiary structure of G-quadruplexes is strongly dependent on the loop sequence which is disregarded by the generally accepted algorithm, we hypothesized that G-quadruplexes with similar topology and, indirectly, similar interaction patterns, can be determined using phylogenetic clustering based on differences in the loop sequences. Phylogenetic analysis of 52 G-quadruplex forming sequences in the Escherichia coli genome revealed two conserved G-quadruplex motifs with a potential regulatory role. Further analysis revealed that both motifs tend to form hairpins and G quadruplexes, as supported by circular dichroism studies. The phylogenetic analysis as described in this work can greatly improve the discovery of functional G-quadruplex structures and may explain unknown regulatory patterns.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
QGRS-Conserve: a computational method for discovering evolutionarily conserved G-quadruplex motifs.Hum Genomics. 2014 May 1;8(1):8. doi: 10.1186/1479-7364-8-8. Hum Genomics. 2014. PMID: 24885782 Free PMC article.
-
Distance-dependent duplex DNA destabilization proximal to G-quadruplex/i-motif sequences.Nucleic Acids Res. 2013 Aug;41(15):7453-61. doi: 10.1093/nar/gkt476. Epub 2013 Jun 14. Nucleic Acids Res. 2013. PMID: 23771141 Free PMC article.
-
The Presence and Localization of G-Quadruplex Forming Sequences in the Domain of Bacteria.Molecules. 2019 May 2;24(9):1711. doi: 10.3390/molecules24091711. Molecules. 2019. PMID: 31052562 Free PMC article.
-
Beyond G-Quadruplexes-The Effect of Junction with Additional Structural Motifs on Aptamers Properties.Int J Mol Sci. 2021 Sep 14;22(18):9948. doi: 10.3390/ijms22189948. Int J Mol Sci. 2021. PMID: 34576112 Free PMC article. Review.
-
DNA and RNA quadruplex-binding proteins.Int J Mol Sci. 2014 Sep 29;15(10):17493-517. doi: 10.3390/ijms151017493. Int J Mol Sci. 2014. PMID: 25268620 Free PMC article. Review.
Cited by
-
Essential Roles and Risks of G-Quadruplex Regulation: Recognition Targets of ALS-Linked TDP-43 and FUS.Front Mol Biosci. 2022 Jul 11;9:957502. doi: 10.3389/fmolb.2022.957502. eCollection 2022. Front Mol Biosci. 2022. PMID: 35898304 Free PMC article. Review.
-
Long Tracts of Guanines Drive Aggregation of RNA G-Quadruplexes in the Presence of Spermine.Biochemistry. 2021 Sep 14;60(36):2715-2726. doi: 10.1021/acs.biochem.1c00467. Epub 2021 Aug 27. Biochemistry. 2021. PMID: 34448586 Free PMC article.
-
G-quadruplexes: a promising target for cancer therapy.Mol Cancer. 2021 Feb 25;20(1):40. doi: 10.1186/s12943-021-01328-4. Mol Cancer. 2021. PMID: 33632214 Free PMC article. Review.
-
Altering translation allows E. coli to overcome chemically stabilized G-quadruplexes.bioRxiv [Preprint]. 2024 Aug 12:2024.08.12.607615. doi: 10.1101/2024.08.12.607615. bioRxiv. 2024. Update in: Nucleic Acids Res. 2025 Mar 20;53(6):gkaf264. doi: 10.1093/nar/gkaf264. PMID: 39185182 Free PMC article. Updated. Preprint.
-
Unraveling the Regulatory G-Quadruplex Puzzle: Lessons From Genome and Transcriptome-Wide Studies.Front Genet. 2019 Oct 18;10:1002. doi: 10.3389/fgene.2019.01002. eCollection 2019. Front Genet. 2019. PMID: 31681431 Free PMC article. Review.
References
-
- Pérez-Martín J., de Lorenzo V. Clues and consequences of DNA bending in transcription. Annu. Rev. Microbiol. 1997;51:593–628. - PubMed
-
- Hatfield G.W., Benham C.J. DNA topology-mediated control of global gene expression in Escherichia coli. Annu. Rev. Genet. 2002;36:175–203. - PubMed
-
- Murat P., Balasubramanian S. Existence and consequences of G-quadruplex structures in DNA. Curr. Opin. Genet. Dev. 2014;25:22–29. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

