The HemQ coprohaem decarboxylase generates reactive oxygen species: implications for the evolution of classical haem biosynthesis
- PMID: 27597779
- PMCID: PMC5095920
- DOI: 10.1042/BCJ20160696
The HemQ coprohaem decarboxylase generates reactive oxygen species: implications for the evolution of classical haem biosynthesis
Abstract
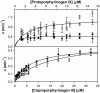
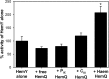
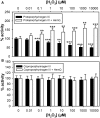
Bacteria require a haem biosynthetic pathway for the assembly of a variety of protein complexes, including cytochromes, peroxidases, globins, and catalase. Haem is synthesised via a series of tetrapyrrole intermediates, including non-metallated porphyrins, such as protoporphyrin IX, which is well known to generate reactive oxygen species in the presence of light and oxygen. Staphylococcus aureus has an ancient haem biosynthetic pathway that proceeds via the formation of coproporphyrin III, a less reactive porphyrin. Here, we demonstrate, for the first time, that HemY of S. aureus is able to generate both protoporphyrin IX and coproporphyrin III, and that the terminal enzyme of this pathway, HemQ, can stimulate the generation of protoporphyrin IX (but not coproporphyrin III). Assays with hydrogen peroxide, horseradish peroxidase, superoxide dismutase, and catalase confirm that this stimulatory effect is mediated by superoxide. Structural modelling reveals that HemQ enzymes do not possess the structural attributes that are common to peroxidases that form compound I [FeIV==O]+, which taken together with the superoxide data leaves Fenton chemistry as a likely route for the superoxide-mediated stimulation of protoporphyrinogen IX oxidase activity of HemY. This generation of toxic free radicals could explain why HemQ enzymes have not been identified in organisms that synthesise haem via the classical protoporphyrin IX pathway. This work has implications for the divergent evolution of haem biosynthesis in ancestral microorganisms, and provides new structural and mechanistic insights into a recently discovered oxidative decarboxylase reaction.
Keywords: coproporphyrinogen; protoporphyrinogen.
© 2016 The Author(s).
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

