Direct Correlation between Motile Behavior and Protein Abundance in Single Cells
- PMID: 27599206
- PMCID: PMC5012591
- DOI: 10.1371/journal.pcbi.1005041
Direct Correlation between Motile Behavior and Protein Abundance in Single Cells
Erratum in
-
Correction: Direct Correlation between Motile Behavior and Protein Abundance in Single Cells.PLoS Comput Biol. 2016 Sep 30;12(9):e1005149. doi: 10.1371/journal.pcbi.1005149. eCollection 2016 Sep. PLoS Comput Biol. 2016. PMID: 27689803 Free PMC article.
Abstract
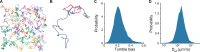
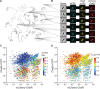
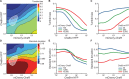
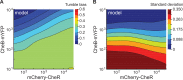
Understanding how stochastic molecular fluctuations affect cell behavior requires the quantification of both behavior and protein numbers in the same cells. Here, we combine automated microscopy with in situ hydrogel polymerization to measure single-cell protein expression after tracking swimming behavior. We characterized the distribution of non-genetic phenotypic diversity in Escherichia coli motility, which affects single-cell exploration. By expressing fluorescently tagged chemotaxis proteins (CheR and CheB) at different levels, we quantitatively mapped motile phenotype (tumble bias) to protein numbers using thousands of single-cell measurements. Our results disagreed with established models until we incorporated the role of CheB in receptor deamidation and the slow fluctuations in receptor methylation. Beyond refining models, our central finding is that changes in numbers of CheR and CheB affect the population mean tumble bias and its variance independently. Therefore, it is possible to adjust the degree of phenotypic diversity of a population by adjusting the global level of expression of CheR and CheB while keeping their ratio constant, which, as shown in previous studies, confers functional robustness to the system. Since genetic control of protein expression is heritable, our results suggest that non-genetic diversity in motile behavior is selectable, supporting earlier hypotheses that such diversity confers a selective advantage.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Ultrasensitivity and fluctuations in the Barkai-Leibler model of chemotaxis receptors in Escherichia coli.PLoS One. 2017 Apr 13;12(4):e0175309. doi: 10.1371/journal.pone.0175309. eCollection 2017. PLoS One. 2017. PMID: 28406996 Free PMC article.
-
CheR- and CheB-dependent chemosensory adaptation system of Rhodobacter sphaeroides.J Bacteriol. 2001 Dec;183(24):7135-44. doi: 10.1128/JB.183.24.7135-7144.2001. J Bacteriol. 2001. PMID: 11717272 Free PMC article.
-
Chemotaxis in Escherichia coli: a molecular model for robust precise adaptation.PLoS Comput Biol. 2008 Jan;4(1):e1. doi: 10.1371/journal.pcbi.0040001. Epub 2007 Nov 20. PLoS Comput Biol. 2008. PMID: 18179279 Free PMC article.
-
Methylation-independent aerotaxis mediated by the Escherichia coli Aer protein.J Bacteriol. 2004 Jun;186(12):3730-7. doi: 10.1128/JB.186.12.3730-3737.2004. J Bacteriol. 2004. PMID: 15175286 Free PMC article.
-
Behavioral Variability and Phenotypic Diversity in Bacterial Chemotaxis.Annu Rev Biophys. 2018 May 20;47:595-616. doi: 10.1146/annurev-biophys-062215-010954. Epub 2018 Apr 4. Annu Rev Biophys. 2018. PMID: 29618219 Free PMC article. Review.
Cited by
-
Non-Genetic Diversity in Chemosensing and Chemotactic Behavior.Int J Mol Sci. 2021 Jun 28;22(13):6960. doi: 10.3390/ijms22136960. Int J Mol Sci. 2021. PMID: 34203411 Free PMC article. Review.
-
MotGen: a closed-loop bacterial motility control framework using generative adversarial networks.Bioinformatics. 2024 Mar 29;40(4):btae170. doi: 10.1093/bioinformatics/btae170. Bioinformatics. 2024. PMID: 38552318 Free PMC article.
-
Alkaline pH Increases Swimming Speed and Facilitates Mucus Penetration for Vibrio cholerae.J Bacteriol. 2021 Mar 8;203(7):e00607-20. doi: 10.1128/JB.00607-20. Print 2021 Mar 8. J Bacteriol. 2021. PMID: 33468594 Free PMC article.
-
Flagellar motor remodeling during swarming requires FliL.bioRxiv [Preprint]. 2023 Jul 14:2023.07.14.549092. doi: 10.1101/2023.07.14.549092. bioRxiv. 2023. Update in: Mol Microbiol. 2023 Nov;120(5):670-683. doi: 10.1111/mmi.15148. PMID: 37503052 Free PMC article. Updated. Preprint.
-
Phenotypic diversity and temporal variability in a bacterial signaling network revealed by single-cell FRET.Elife. 2017 Dec 12;6:e27455. doi: 10.7554/eLife.27455. Elife. 2017. PMID: 29231170 Free PMC article.
References
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

