Functionally identifiable apoptosis-insensitive subpopulations determine chemoresistance in acute myeloid leukemia
- PMID: 27599292
- PMCID: PMC5096802
- DOI: 10.1172/JCI82908
Functionally identifiable apoptosis-insensitive subpopulations determine chemoresistance in acute myeloid leukemia
Abstract
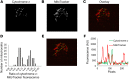
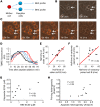
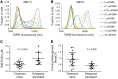
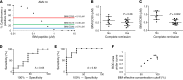
Upfront resistance to chemotherapy and relapse following remission are critical problems in leukemia that are generally attributed to subpopulations of chemoresistant tumor cells. There are, however, limited means for prospectively identifying these subpopulations, which hinders an understanding of therapeutic resistance. BH3 profiling is a functional single-cell analysis using synthetic BCL-2 BH3 domain-like peptides that measures mitochondrial apoptotic sensitivity or "priming." Here, we observed that the extent of apoptotic priming is heterogeneous within multiple cancer cell lines and is not the result of experimental noise. Apoptotic priming was also heterogeneous in treatment-naive primary human acute myeloid leukemia (AML) myeloblasts, and this heterogeneity decreased in chemotherapy-treated AML patients. The priming of the most apoptosis-resistant tumor cells, rather than the median priming of the population, best predicted patient response to induction chemotherapy. For several patients, these poorly primed subpopulations of AML tumor cells were enriched for antiapoptotic proteins. Developing techniques to identify and understand these apoptosis-insensitive subpopulations of tumor cells may yield insights into clinical chemoresistance and potentially improve therapeutic outcomes in AML.
Figures


References
-
- Keating MJ, et al. Response to salvage therapy and survival after relapse in acute myelogenous leukemia. J Clin Oncol. 1989;7(8):1071–1080. - PubMed
-
- Kantarjian HM, Keating MJ, Walters RS, McCredie KB, Freireich EJ. The characteristics and outcome of patients with late relapse acute myelogenous leukemia. J Clin Oncol. 1988;6(2):232–238. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

