TGF-β receptor maintains CD4 T helper cell identity during chronic viral infections
- PMID: 27599295
- PMCID: PMC5096797
- DOI: 10.1172/JCI87041
TGF-β receptor maintains CD4 T helper cell identity during chronic viral infections
Abstract
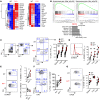
Suppression of CD8 and CD4 T cells is a hallmark in chronic viral infections, including hepatitis C and HIV. While multiple pathways are known to inhibit CD8 T cells, the host molecules that restrict CD4 T cell responses are less understood. Here, we used inducible and CD4 T cell-specific deletion of the gene encoding the TGF-β receptor during chronic lymphocytic choriomeningitis virus infection in mice, and determined that TGF-β signaling restricted proliferation and terminal differentiation of antiviral CD4 T cells. TGF-β signaling also inhibited a cytotoxic program that includes granzymes and perforin expression at both early and late stages of infection in vivo and repressed the transcription factor eomesodermin. Overexpression of eomesodermin was sufficient to recapitulate in great part the phenotype of TGF-β receptor-deficient CD4 T cells, while SMAD4 was necessary for CD4 T cell accumulation and differentiation. TGF-β signaling also restricted accumulation and differentiation of CD4 T cells and reduced the expression of cytotoxic molecules in mice and humans infected with other persistent viruses. These data uncovered an eomesodermin-driven CD4 T cell program that is continuously suppressed by TGF-β signaling. During chronic viral infection, this program limits CD4 T cell responses while maintaining CD4 T helper cell identity.
Figures





Similar articles
-
Ablation of CD8 and CD4 T cell responses by high viral loads.J Immunol. 2003 Jan 1;170(1):477-86. doi: 10.4049/jimmunol.170.1.477. J Immunol. 2003. PMID: 12496434
-
Homeostatic turnover of virus-specific memory CD8 T cells occurs stochastically and is independent of CD4 T cell help.J Immunol. 2010 Sep 15;185(6):3436-44. doi: 10.4049/jimmunol.1001421. Epub 2010 Aug 23. J Immunol. 2010. PMID: 20733203
-
TGF-β blockade does not improve control of an established persistent viral infection.Viral Immunol. 2012 Jun;25(3):232-8. doi: 10.1089/vim.2011.0079. Epub 2012 May 23. Viral Immunol. 2012. PMID: 22620718 Free PMC article.
-
Chronic LCMV Infection Is Fortified with Versatile Tactics to Suppress Host T Cell Immunity and Establish Viral Persistence.Viruses. 2021 Sep 29;13(10):1951. doi: 10.3390/v13101951. Viruses. 2021. PMID: 34696381 Free PMC article. Review.
-
Fighting Viral Infections and Virus-Driven Tumors with Cytotoxic CD4+ T Cells.Front Immunol. 2017 Feb 27;8:197. doi: 10.3389/fimmu.2017.00197. eCollection 2017. Front Immunol. 2017. PMID: 28289418 Free PMC article. Review.
Cited by
-
TGF-β regulates the stem-like state of PD-1+ TCF-1+ virus-specific CD8 T cells during chronic infection.J Exp Med. 2022 Oct 3;219(10):e20211574. doi: 10.1084/jem.20211574. Epub 2022 Aug 18. J Exp Med. 2022. PMID: 35980386 Free PMC article.
-
CD4+ T Cell Differentiation in Chronic Viral Infections: The Tfh Perspective.Trends Mol Med. 2017 Dec;23(12):1072-1087. doi: 10.1016/j.molmed.2017.10.001. Epub 2017 Nov 12. Trends Mol Med. 2017. PMID: 29137933 Free PMC article. Review.
-
SKI and SMAD4 are essential for IL-21-induced Th17 differentiation.Mol Immunol. 2019 Oct;114:260-268. doi: 10.1016/j.molimm.2019.07.029. Epub 2019 Aug 6. Mol Immunol. 2019. PMID: 31398665 Free PMC article.
-
Consequences of Mutations and Abnormal Expression of SMAD4 in Tumors and T Cells.Onco Targets Ther. 2021 Apr 13;14:2531-2540. doi: 10.2147/OTT.S297855. eCollection 2021. Onco Targets Ther. 2021. PMID: 33888990 Free PMC article. Review.
-
TGF-β in T Cell Biology: Implications for Cancer Immunotherapy.Cancers (Basel). 2018 Jun 11;10(6):194. doi: 10.3390/cancers10060194. Cancers (Basel). 2018. PMID: 29891791 Free PMC article. Review.
References
-
- Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12(6):492–499. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

