Genome-wide analyses of long noncoding RNA expression profiles correlated with radioresistance in nasopharyngeal carcinoma via next-generation deep sequencing
- PMID: 27599611
- PMCID: PMC5012053
- DOI: 10.1186/s12885-016-2755-6
Genome-wide analyses of long noncoding RNA expression profiles correlated with radioresistance in nasopharyngeal carcinoma via next-generation deep sequencing
Abstract
Background: Radioresistance is one of the major factors limiting the therapeutic efficacy and prognosis of patients with nasopharyngeal carcinoma (NPC). Accumulating evidence has suggested that aberrant expression of long noncoding RNAs (lncRNAs) contributes to cancer progression. Therefore, here we identified lncRNAs associated with radioresistance in NPC.
Methods: The differential expression profiles of lncRNAs associated with NPC radioresistance were constructed by next-generation deep sequencing by comparing radioresistant NPC cells with their parental cells. LncRNA-related mRNAs were predicted and analyzed using bioinformatics algorithms compared with the mRNA profiles related to radioresistance obtained in our previous study. Several lncRNAs and associated mRNAs were validated in established NPC radioresistant cell models and NPC tissues.
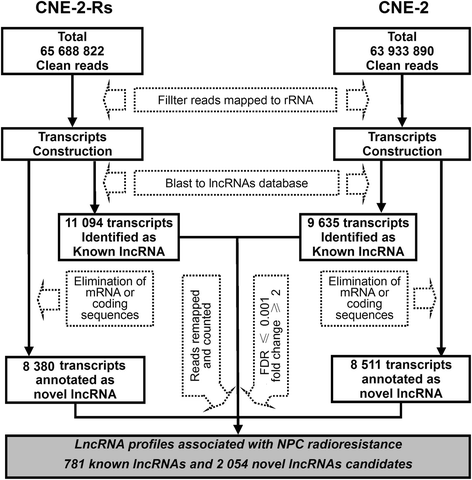
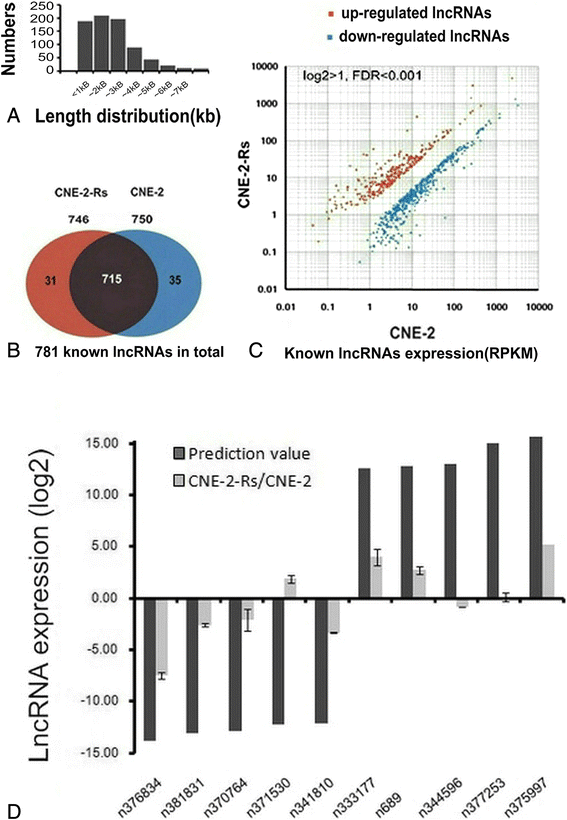
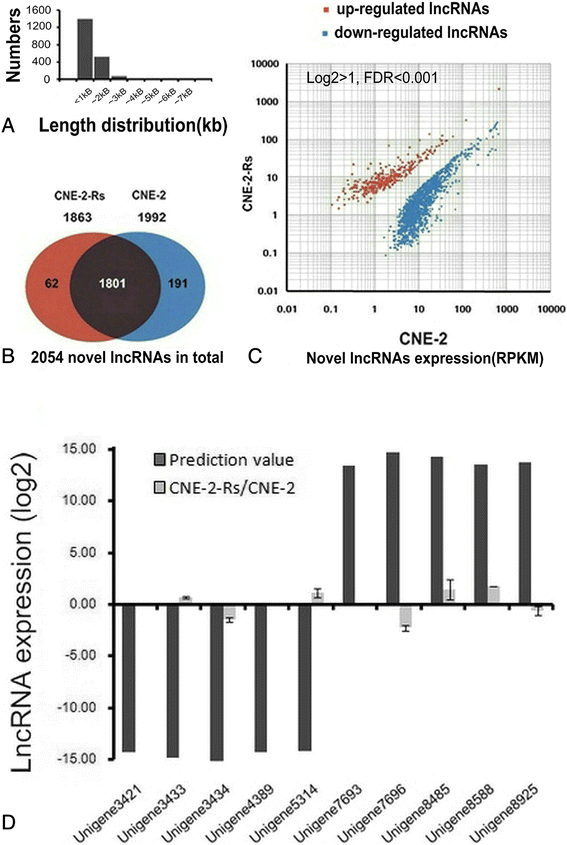
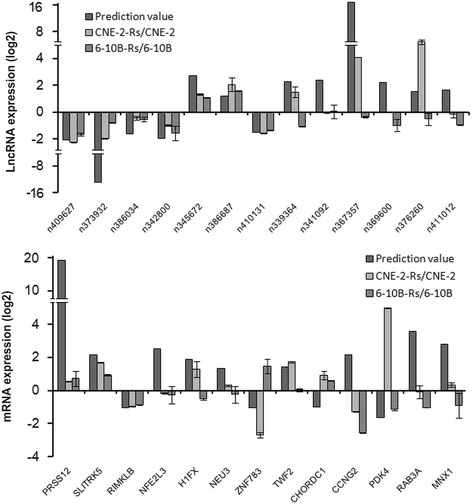
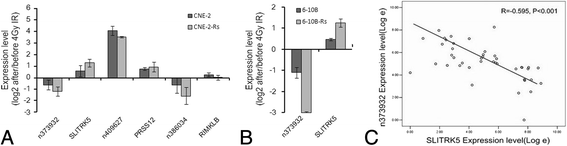
Results: By comparison between radioresistant CNE-2-Rs and parental CNE-2 cells by next-generation deep sequencing, a total of 781 known lncRNAs and 2054 novel lncRNAs were annotated. The top five upregulated and downregulated known/novel lncRNAs were detected using quantitative real-time reverse transcription-polymerase chain reaction, and 7/10 known lncRNAs and 3/10 novel lncRNAs were demonstrated to have significant differential expression trends that were the same as those predicted by deep sequencing. From the prediction process, 13 pairs of lncRNAs and their associated genes were acquired, and the prediction trends of three pairs were validated in both radioresistant CNE-2-Rs and 6-10B-Rs cell lines, including lncRNA n373932 and SLITRK5, n409627 and PRSS12, and n386034 and RIMKLB. LncRNA n373932 and its related SLITRK5 showed dramatic expression changes in post-irradiation radioresistant cells and a negative expression correlation in NPC tissues (R = -0.595, p < 0.05).
Conclusions: Our study provides an overview of the expression profiles of radioresistant lncRNAs and potentially related mRNAs, which will facilitate future investigations into the function of lncRNAs in NPC radioresistance.
Keywords: Deep sequencing; Long noncoding RNA; Nasopharyngeal carcinoma; Radioresistance.
Figures
Similar articles
-
Next generation deep sequencing identified a novel lncRNA n375709 associated with paclitaxel resistance in nasopharyngeal carcinoma.Oncol Rep. 2016 Oct;36(4):1861-7. doi: 10.3892/or.2016.4981. Epub 2016 Jul 28. Oncol Rep. 2016. PMID: 27498905 Free PMC article.
-
An integrative transcriptomic analysis reveals p53 regulated miRNA, mRNA, and lncRNA networks in nasopharyngeal carcinoma.Tumour Biol. 2016 Mar;37(3):3683-95. doi: 10.1007/s13277-015-4156-x. Epub 2015 Oct 13. Tumour Biol. 2016. PMID: 26462838
-
LncRNA CASC19 Enhances the Radioresistance of Nasopharyngeal Carcinoma by Regulating the miR-340-3p/FKBP5 Axis.Int J Mol Sci. 2023 Feb 3;24(3):3047. doi: 10.3390/ijms24033047. Int J Mol Sci. 2023. PMID: 36769373 Free PMC article.
-
Functions and Roles of Long-Non-Coding RNAs in Human Nasopharyngeal Carcinoma.Cell Physiol Biochem. 2018;45(3):1191-1204. doi: 10.1159/000487451. Epub 2018 Feb 9. Cell Physiol Biochem. 2018. PMID: 29448252 Review.
-
Radio-Susceptibility of Nasopharyngeal Carcinoma: Focus on Epstein- Barr Virus, MicroRNAs, Long Non-Coding RNAs and Circular RNAs.Curr Mol Pharmacol. 2020;13(3):192-205. doi: 10.2174/1874467213666191227104646. Curr Mol Pharmacol. 2020. PMID: 31880267 Review.
Cited by
-
RNA-Seq analysis of peripheral blood mononuclear cells reveals unique transcriptional signatures associated with radiotherapy response of nasopharyngeal carcinoma and prognosis of head and neck cancer.Cancer Biol Ther. 2020;21(2):139-146. doi: 10.1080/15384047.2019.1670521. Epub 2019 Nov 7. Cancer Biol Ther. 2020. PMID: 31698994 Free PMC article.
-
USP44 regulates irradiation-induced DNA double-strand break repair and suppresses tumorigenesis in nasopharyngeal carcinoma.Nat Commun. 2022 Jan 25;13(1):501. doi: 10.1038/s41467-022-28158-2. Nat Commun. 2022. PMID: 35079021 Free PMC article.
-
Comprehensive analysis of key genes and microRNAs in radioresistant nasopharyngeal carcinoma.BMC Med Genomics. 2019 May 28;12(1):73. doi: 10.1186/s12920-019-0507-6. BMC Med Genomics. 2019. PMID: 31138194 Free PMC article.
-
Overexpression of N-cadherin and β-catenin correlates with poor prognosis in patients with nasopharyngeal carcinoma.Oncol Lett. 2017 Mar;13(3):1725-1730. doi: 10.3892/ol.2017.5645. Epub 2017 Jan 25. Oncol Lett. 2017. PMID: 28454316 Free PMC article.
-
Long non-coding RNA NEAT1 regulates epithelial membrane protein 2 expression to repress nasopharyngeal carcinoma migration and irradiation-resistance through miR-101-3p as a competing endogenous RNA mechanism.Oncotarget. 2017 Jul 26;8(41):70156-70171. doi: 10.18632/oncotarget.19596. eCollection 2017 Sep 19. Oncotarget. 2017. PMID: 29050268 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

