Reactivation of mutant p53 by capsaicin, the major constituent of peppers
- PMID: 27599722
- PMCID: PMC5012067
- DOI: 10.1186/s13046-016-0417-9
Reactivation of mutant p53 by capsaicin, the major constituent of peppers
Abstract
Background: Mutations in the p53 oncosuppressor gene are highly frequent in human cancers. These alterations are mainly point mutations in the DNA binding domain of p53 and disable p53 from transactivating target genes devoted to anticancer activity. Mutant p53 proteins are usually more stable than wild-type p53 and may not only impair wild-type p53 activity but also acquire pro-oncogenic functions. Therefore, targeting mutant p53 to clear the hyperstable proteins or change p53 conformation to reactivate wild-type p53 protein functions is a powerful anticancer strategy. Several small molecules have been tested for p53 reactivation in mutant p53-carrying cells while studies exploiting the effect of natural compounds are limited. Capsaicin (CPS) is the major constituent of peppers and show antitumor activity by targeting several molecular pathway, however, its effect on mutant p53 reactivation has not been assessed yet. In this study we aimed at investigating whether mutant p53 could be a new target of capsaicin-induced cell death and the underlying mechanisms.
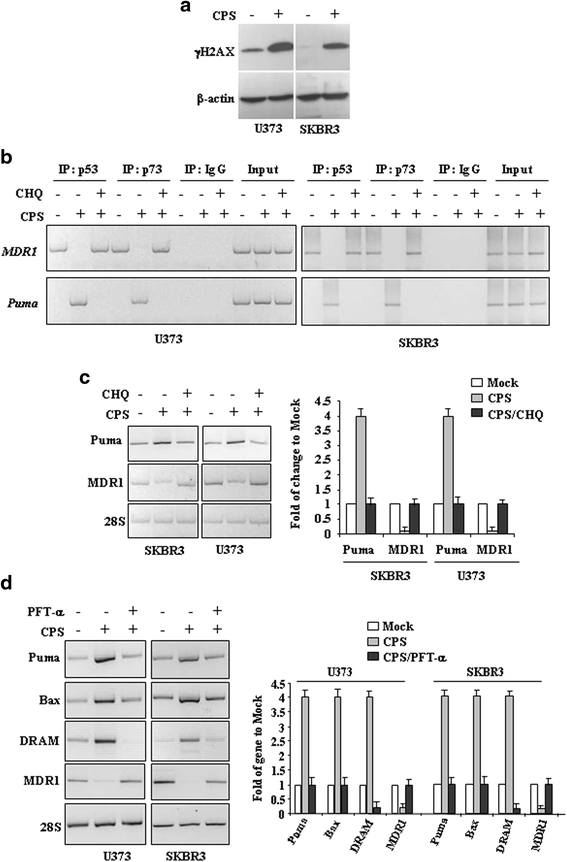
Methods: p53 levels were analysed by western blot upon capsaicin treatment in the presence of the autophagy inhibitor chloroquine. The mutant p53 reactivation was evaluated by chromatin-immunoprecipitation (ChIP) assay and semi-quantitative RT-PCR analyses of wild-type p53 target genes. The specific wild-type p53 activation was determined by using the inhibitor of p53 transactivation function, pifithrin-α and siRNA for p53.
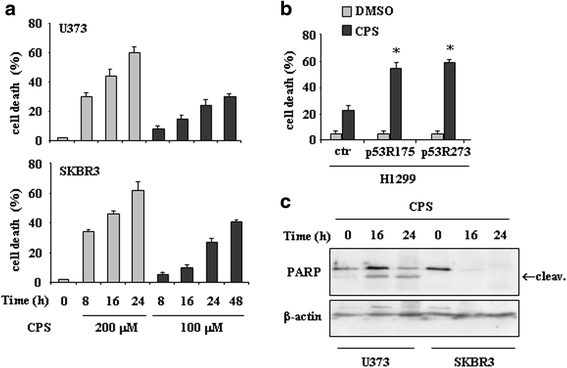
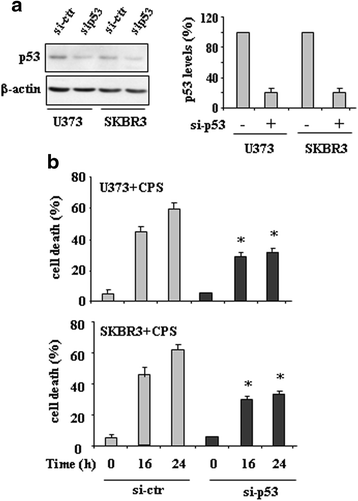
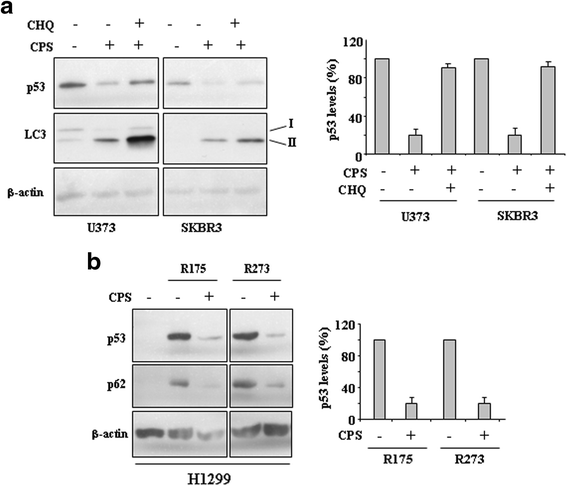
Results: Here, we show that capsaicin induced autophagy that was, at least in part, responsible of mutant p53 protein degradation. Abrogation of mutant p53 by capsaicin restored wild-type p53 activities over mutant p53 functions, contributing to cancer cell death. Similar effects were confirmed in cancer cells bearing tumor-associated p53 mutations and in H1299 (p53 null) with overexpressed p53R175H and p53R273H mutant proteins.
Conclusion: These findings demonstrate for the first time that capsaicin may reduce mutant p53 levels and reactivate wild-type p53 protein in mutant p53-carrying cells and the p53 reactivation contributes to capsaicin-induced cell death.
Keywords: Apoptosis; Autophagy; Capsaicin; Mutant p53; Natural compounds; p53; p53 reactivation.
Figures
Similar articles
-
p53 mutant-type in human prostate cancer cells determines the sensitivity to phenethyl isothiocyanate induced growth inhibition.J Exp Clin Cancer Res. 2019 Jul 15;38(1):307. doi: 10.1186/s13046-019-1267-z. J Exp Clin Cancer Res. 2019. PMID: 31307507 Free PMC article.
-
Reactivation of mutant p53 by a dietary-related compound phenethyl isothiocyanate inhibits tumor growth.Cell Death Differ. 2016 Oct;23(10):1615-27. doi: 10.1038/cdd.2016.48. Epub 2016 Jun 3. Cell Death Differ. 2016. PMID: 27258787 Free PMC article.
-
TAR1, a human anti-p53 single-chain antibody, restores tumor suppressor function to mutant p53 variants.J Immunother. 2010 Feb-Mar;33(2):146-54. doi: 10.1097/CJI.0b013e3181be14dc. J Immunother. 2010. PMID: 20139776
-
Anticancer Properties of Capsaicin Against Human Cancer.Anticancer Res. 2016 Mar;36(3):837-43. Anticancer Res. 2016. PMID: 26976969 Review.
-
New therapeutic strategies to treat human cancers expressing mutant p53 proteins.J Exp Clin Cancer Res. 2018 Feb 15;37(1):30. doi: 10.1186/s13046-018-0705-7. J Exp Clin Cancer Res. 2018. PMID: 29448954 Free PMC article. Review.
Cited by
-
Ginsenoside Rh4 Inhibits Colorectal Cancer Cell Proliferation by Inducing Ferroptosis via Autophagy Activation.Evid Based Complement Alternat Med. 2022 May 29;2022:6177553. doi: 10.1155/2022/6177553. eCollection 2022. Evid Based Complement Alternat Med. 2022. PMID: 35677385 Free PMC article.
-
The impact of capsaicinoids on APP processing in Alzheimer's disease in SH-SY5Y cells.Sci Rep. 2020 Jun 8;10(1):9164. doi: 10.1038/s41598-020-66009-6. Sci Rep. 2020. PMID: 32514053 Free PMC article.
-
The deubiquitinase inhibitor WP1130 drives nuclear aggregation and reactivation of mutant p53 for selective cancer cell targeting.FEBS J. 2025 Jun;292(11):2823-2842. doi: 10.1111/febs.70036. Epub 2025 Mar 11. FEBS J. 2025. PMID: 40070163 Free PMC article.
-
Mutant p53 in cancer: from molecular mechanism to therapeutic modulation.Cell Death Dis. 2022 Nov 18;13(11):974. doi: 10.1038/s41419-022-05408-1. Cell Death Dis. 2022. PMID: 36400749 Free PMC article. Review.
-
Mutant p53 and Cellular Stress Pathways: A Criminal Alliance That Promotes Cancer Progression.Cancers (Basel). 2019 May 2;11(5):614. doi: 10.3390/cancers11050614. Cancers (Basel). 2019. PMID: 31052524 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

