Establishment of a simple and efficient Agrobacterium-mediated transformation system for Phytophthora palmivora
- PMID: 27599726
- PMCID: PMC5012004
- DOI: 10.1186/s12866-016-0825-1
Establishment of a simple and efficient Agrobacterium-mediated transformation system for Phytophthora palmivora
Abstract
Background: As an agriculturally important oomycete genus, Phytophthora contains a large number of destructive plant pathogens that severely threaten agricultural production and natural ecosystems. Among them is the broad host range pathogen P. palmivora, which infects many economically important plant species. An essential way to dissect their pathogenesis mechanisms is genetic modification of candidate genes, which requires effective transformation systems. Four methods were developed for transformation of Phytophthora spp., including PEG(polyethylene glycol)/CaCl2 mediated protoplast transformation, electroporation of zoospores, microprojectile bombardment and Agrobacterium-mediated transformation (AMT). Among them, AMT has many advantages over the other methods such as easy handling and mainly generating single-copy integration in the genome. An AMT method previously reported for P. infestans and P. palmivora has barely been used in oomycete research due to low success and low reproducibility.
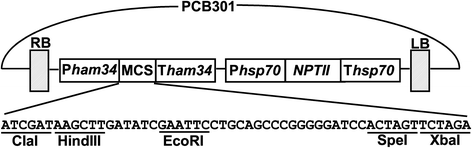
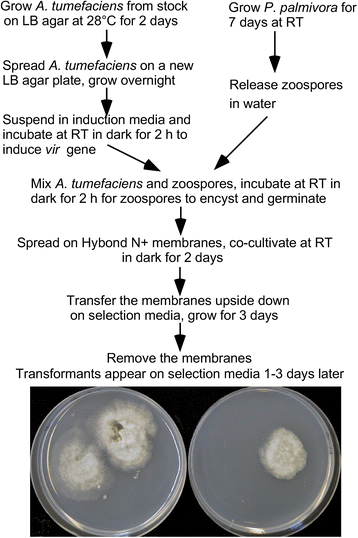
Results: In this study, we report a simple and efficient AMT system for P. palmivora. Using this system, we were able to reproducibly generate over 40 transformants using zoospores collected from culture grown in a single 100 mm-diameter petri dish. The generated GFP transformants constitutively expressed GFP readily detectable using a fluorescence microscope. All of the transformants tested using Southern blot analysis contained a single-copy T-DNA insertion.
Conclusions: This system is highly effective and reproducible for transformation of P. palmivora and expected to be adaptable for transformation of additional Phytophthora spp. and other oomycetes. Its establishment will greatly accelerate their functional genomic studies.
Keywords: Agrobacterium-mediated transformation; Copy number; GFP; Oomycete; Phytophthora palmivora.
Figures
References
-
- Margulis L, Schwartz KV. Five Kingdoms: An Illustrated Guide to the Phyla of Life on Earth. New York: Freeman; 2000.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

