Role of GATA transcription factor ELT-2 and p38 MAPK PMK-1 in recovery from acute P. aeruginosa infection in C. elegans
- PMID: 27600703
- PMCID: PMC5411242
- DOI: 10.1080/21505594.2016.1222334
Role of GATA transcription factor ELT-2 and p38 MAPK PMK-1 in recovery from acute P. aeruginosa infection in C. elegans
Abstract
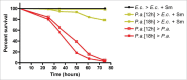
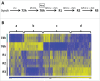
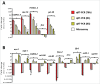
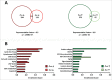
Infectious diseases caused by bacterial pathogens reduce the fitness of their associated host but are generally limited in duration. In order for the diseased host to regain any lost fitness upon recovery, a variety of molecular, cellular, and physiological processes must be employed. To better understand mechanisms underlying the recovery process, we have modeled an acute Pseudomonas aeruginosa infection in C. elegans using brief exposures to this pathogen and subsequent antibiotic treatment. To identify host genes altered during recovery from P. aeruginosa infection, we performed whole genome expression profiling. The analysis of this dataset indicated that the activity of the host immune system is down-regulated upon recovery and revealed shared and pathogen-specific host responses during recovery. We determined that the GATA transcription factor ELT-2 and the p38 MAP kinase PMK-1 are necessary for animals to successfully recover from an acute P. aeruginosa infection. In addition, we found that ELT-2 plays a more prominent and earlier role than PMK-1 during recovery. Our data sheds further light on the molecular mechanisms and transcriptional programs involved in recovery from an acute bacterial infection, which provides a better understanding of the entire infectious disease process.
Keywords: C. elegans; ELT-2; GATA; P. aeruginosa; PMK-1; acute; disease; host; immunity; infection; infectious; innate; p38 MAPK; pathogen; recovery; resolution.
Figures
Comment in
-
Expanding the nematode model system: The molecular basis of inflammation and infection recovery in C. elegans.Virulence. 2017 Apr 3;8(3):244-245. doi: 10.1080/21505594.2016.1239011. Epub 2016 Sep 26. Virulence. 2017. PMID: 27668316 Free PMC article. No abstract available.
References
-
- Buckley CD, Gilroy DW, Serhan CN, Stockinger B, Tak PP. The resolution of inflammation. Nat Rev Immunol 2013; 13:59-66; PMID:23197111; http://dx.doi.org/10.1038/nri3362 - DOI - PubMed
-
- Buckley CD, Gilroy DW, Serhan CN. Proresolving lipid mediators and mechanisms in the resolution of acute inflammation. Immunity 2014; 40:315-27; PMID:24656045; http://dx.doi.org/10.1016/j.immuni.2014.02.009 - DOI - PMC - PubMed
-
- Serhan CN. Pro-resolving lipid mediators are leads for resolution physiology. Nature 2014; 510:92-101. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24899309; PMID:24899309; http://dx.doi.org/10.1038/nature13479 - DOI - PMC - PubMed
-
- Head B, Aballay A. Recovery from an acute infection in C. elegans requires the GATA transcription factor ELT-2. PLoS Genet 2014; 10:e1004609; PMID:25340560; http://dx.doi.org/10.1371/journal.pgen.1004609 - DOI - PMC - PubMed
-
- Torres BY, Oliveira JHM, Thomas Tate A, Rath P, Cumnock K, Schneider DS. Tracking Resilience to Infections by Mapping Disease Space. PLoS Biol 2016; 14:e1002436; PMID:27088359; http://dx.doi.org/10.1371/journal.pbio.1002436 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
