FtsZ-Dependent Elongation of a Coccoid Bacterium
- PMID: 27601570
- PMCID: PMC5013293
- DOI: 10.1128/mBio.00908-16
FtsZ-Dependent Elongation of a Coccoid Bacterium
Abstract
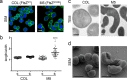
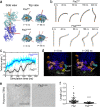
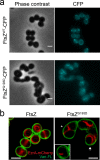
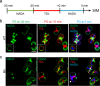
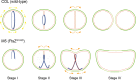
A mechanistic understanding of the determination and maintenance of the simplest bacterial cell shape, a sphere, remains elusive compared with that of more complex shapes. Cocci seem to lack a dedicated elongation machinery, and a spherical shape has been considered an evolutionary dead-end morphology, as a transition from a spherical to a rod-like shape has never been observed in bacteria. Here we show that a Staphylococcus aureus mutant (M5) expressing the ftsZ(G193D) allele exhibits elongated cells. Molecular dynamics simulations and in vitro studies indicate that FtsZ(G193D) filaments are more twisted and shorter than wild-type filaments. In vivo, M5 cell wall deposition is initiated asymmetrically, only on one side of the cell, and progresses into a helical pattern rather than into a constricting ring as in wild-type cells. This helical pattern of wall insertion leads to elongation, as in rod-shaped cells. Thus, structural flexibility of FtsZ filaments can result in an FtsZ-dependent mechanism for generating elongated cells from cocci.
Importance: The mechanisms by which bacteria generate and maintain even the simplest cell shape remain an elusive but fundamental question in microbiology. In the absence of examples of coccus-to-rod transitions, the spherical shape has been suggested to be an evolutionary dead end in morphogenesis. We describe the first observation of the generation of elongated cells from truly spherical cocci, occurring in a Staphylococcus aureus mutant containing a single point mutation in its genome, in the gene encoding the bacterial tubulin homologue FtsZ. We demonstrate that FtsZ-dependent cell elongation is possible, even in the absence of dedicated elongation machinery.
Copyright © 2016 Pereira et al.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
