SCAP/SREBP pathway is required for the full steroidogenic response to cyclic AMP
- PMID: 27601673
- PMCID: PMC5035853
- DOI: 10.1073/pnas.1611424113
SCAP/SREBP pathway is required for the full steroidogenic response to cyclic AMP
Abstract
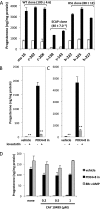
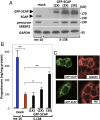
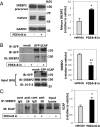
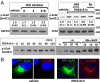
Luteinizing hormone (LH) stimulates steroidogenesis largely through a surge in cyclic AMP (cAMP). Steroidogenic rates are also critically dependent on the availability of cholesterol at mitochondrial sites of synthesis. This cholesterol is provided by cellular uptake of lipoproteins, mobilization of intracellular lipid, and de novo synthesis. Whether and how these pathways are coordinated by cAMP are poorly understood. Recent phosphoproteomic analyses of cAMP-dependent phosphorylation sites in MA10 Leydig cells suggested that cAMP regulates multiple steps in these processes, including activation of the SCAP/SREBP pathway. SCAP [sterol-regulatory element-binding protein (SREBP) cleavage-activating protein] acts as a cholesterol sensor responsible for regulating intracellular cholesterol balance. Its role in cAMP-mediated control of steroidogenesis has not been explored. We used two CRISPR (clustered regularly interspaced short palindromic repeat)-Cas9 (CRISPR associated protein 9) knockout approaches to test the role of SCAP in steroidogenesis. Our results demonstrate that SCAP is required for progesterone production induced by concurrent inhibition of the cAMP phosphodiesterases PDE4 and PDE8. These inhibitors increased SCAP phosphorylation, SREBP2 activation, and subsequent expression of cholesterol biosynthetic genes, whereas SCAP deficiency largely prevented these effects. Reexpression of SCAP in SCAP-deficient cells restored SREBP2 protein expression and partially restored steroidogenic responses, confirming the requirement of SCAP-SREBP2 in steroidogenesis. Inhibitors of 3-hydroxy-3-methylglutaryl-Coenzyme A reductase and isoprenylation attenuated, whereas exogenously provided cholesterol augmented, PDE inhibitor-induced steroidogenesis, suggesting that the cholesterol substrate needed for steroidogenesis is provided by both de novo synthesis and isoprenylation-dependent mechanisms. Overall, these results demonstrate a novel role for LH/cAMP in SCAP/SREBP activation and subsequent regulation of steroidogenesis.
Keywords: SCAP/SREBP; cAMP; cholesterol; phosphodiesterase; steroidogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Bender AT, Beavo JA. Cyclic nucleotide phosphodiesterases: Molecular regulation to clinical use. Pharmacol Rev. 2006;58(3):488–520. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

