Evaluation of 16S rRNA Gene Primer Pairs for Monitoring Microbial Community Structures Showed High Reproducibility within and Low Comparability between Datasets Generated with Multiple Archaeal and Bacterial Primer Pairs
- PMID: 27602022
- PMCID: PMC4994424
- DOI: 10.3389/fmicb.2016.01297
Evaluation of 16S rRNA Gene Primer Pairs for Monitoring Microbial Community Structures Showed High Reproducibility within and Low Comparability between Datasets Generated with Multiple Archaeal and Bacterial Primer Pairs
Abstract
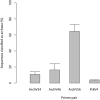
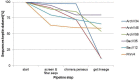
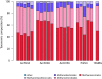
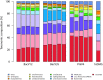
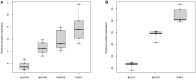
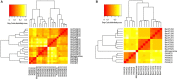
The application of next-generation sequencing technology in microbial community analysis increased our knowledge and understanding of the complexity and diversity of a variety of ecosystems. In contrast to Bacteria, the archaeal domain was often not particularly addressed in the analysis of microbial communities. Consequently, established primers specifically amplifying the archaeal 16S ribosomal gene region are scarce compared to the variety of primers targeting bacterial sequences. In this study, we aimed to validate archaeal primers suitable for high throughput next generation sequencing. Three archaeal 16S primer pairs as well as two bacterial and one general microbial 16S primer pairs were comprehensively tested by in-silico evaluation and performing an experimental analysis of a complex microbial community of a biogas reactor. The results obtained clearly demonstrate that comparability of community profiles established using different primer pairs is difficult. 16S rRNA gene data derived from a shotgun metagenome of the same reactor sample added an additional perspective on the community structure. Furthermore, in-silico evaluation of primers, especially those for amplification of archaeal 16S rRNA gene regions, does not necessarily reflect the results obtained in experimental approaches. In the latter, archaeal primer pair ArchV34 showed the highest similarity to the archaeal community structure compared to observed by the metagenomic approach and thus appears to be the appropriate for analyzing archaeal communities in biogas reactors. However, a disadvantage of this primer pair was its low specificity for the archaeal domain in the experimental application leading to high amounts of bacterial sequences within the dataset. Overall our results indicate a rather limited comparability between community structures investigated and determined using different primer pairs as well as between metagenome and 16S rRNA gene amplicon based community structure analysis. This finding, previously shown for Bacteria, was as well observed for the archaeal domain.
Keywords: 16S microbial community; amplicon sequencing; archaea; metagenome; microbial communities; next-generation sequencing.
Figures

Similar articles
-
Novel primers for 16S rRNA gene-based archaeal and bacterial community analysis in oceanic trench sediments.Appl Microbiol Biotechnol. 2022 Apr;106(7):2795-2809. doi: 10.1007/s00253-022-11893-3. Epub 2022 Mar 29. Appl Microbiol Biotechnol. 2022. PMID: 35348850
-
Fine-scale evaluation of two standard 16S rRNA gene amplicon primer pairs for analysis of total prokaryotes and archaeal nitrifiers in differently managed soils.Front Microbiol. 2023 Feb 23;14:1140487. doi: 10.3389/fmicb.2023.1140487. eCollection 2023. Front Microbiol. 2023. PMID: 36910167 Free PMC article.
-
In-Silico Detection of Oral Prokaryotic Species With Highly Similar 16S rRNA Sequence Segments Using Different Primer Pairs.Front Cell Infect Microbiol. 2022 Feb 9;11:770668. doi: 10.3389/fcimb.2021.770668. eCollection 2021. Front Cell Infect Microbiol. 2022. PMID: 35223533 Free PMC article.
-
Review and re-analysis of domain-specific 16S primers.J Microbiol Methods. 2003 Dec;55(3):541-55. doi: 10.1016/j.mimet.2003.08.009. J Microbiol Methods. 2003. PMID: 14607398 Review.
-
Current status and future developments of assessing microbiome composition and dynamics in anaerobic digestion systems using metagenomic approaches.Heliyon. 2024 Mar 20;10(6):e28221. doi: 10.1016/j.heliyon.2024.e28221. eCollection 2024 Mar 30. Heliyon. 2024. PMID: 38560681 Free PMC article. Review.
Cited by
-
Microbial diversity and ecological roles of halophilic microorganisms in Dingbian (Shaanxi, China) saline-alkali soils and salt lakes.BMC Microbiol. 2025 May 11;25(1):287. doi: 10.1186/s12866-025-03997-3. BMC Microbiol. 2025. PMID: 40350492 Free PMC article.
-
Insights into microbial community structure and diversity in oil palm waste compost.3 Biotech. 2019 Oct;9(10):364. doi: 10.1007/s13205-019-1892-4. Epub 2019 Sep 20. 3 Biotech. 2019. PMID: 31588388 Free PMC article.
-
Primer, Pipelines, Parameters: Issues in 16S rRNA Gene Sequencing.mSphere. 2021 Feb 24;6(1):e01202-20. doi: 10.1128/mSphere.01202-20. mSphere. 2021. PMID: 33627512 Free PMC article.
-
Gut Microbiota beyond Bacteria-Mycobiome, Virome, Archaeome, and Eukaryotic Parasites in IBD.Int J Mol Sci. 2020 Apr 11;21(8):2668. doi: 10.3390/ijms21082668. Int J Mol Sci. 2020. PMID: 32290414 Free PMC article. Review.
-
Evaluating Established Methods for Rumen 16S rRNA Amplicon Sequencing With Mock Microbial Populations.Front Microbiol. 2018 Jun 25;9:1365. doi: 10.3389/fmicb.2018.01365. eCollection 2018. Front Microbiol. 2018. PMID: 29988486 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

