DNA methylation: conducting the orchestra from exposure to phenotype?
- PMID: 27602172
- PMCID: PMC5012062
- DOI: 10.1186/s13148-016-0256-8
DNA methylation: conducting the orchestra from exposure to phenotype?
Abstract
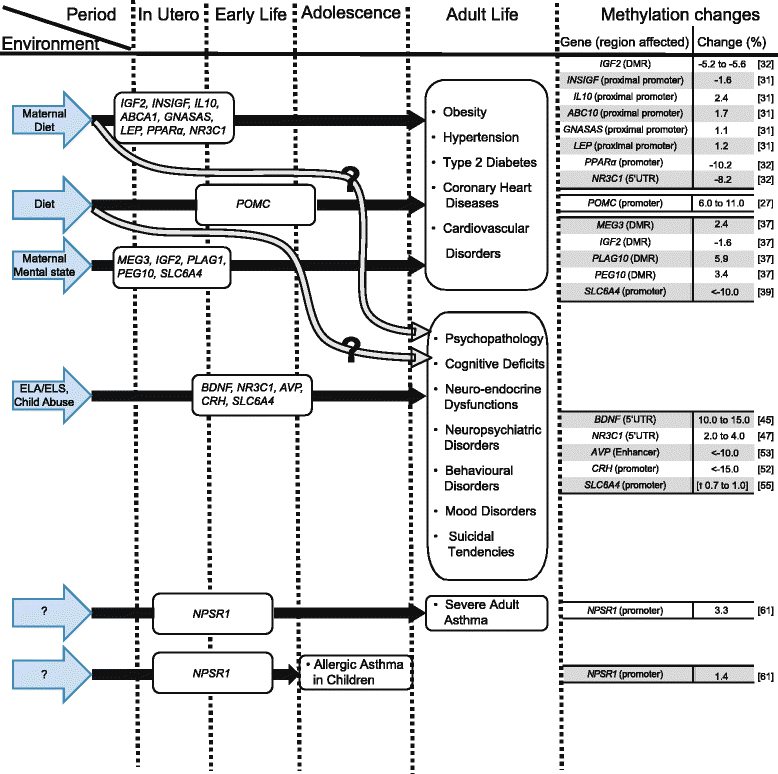
DNA methylation, through 5-methyl- and 5-hydroxymethylcytosine (5mC and 5hmC), is considered to be one of the principal interfaces between the genome and our environment, and it helps explain phenotypic variations in human populations. Initial reports of large differences in methylation level in genomic regulatory regions, coupled with clear gene expression data in both imprinted genes and malignant diseases, provided easily dissected molecular mechanisms for switching genes on or off. However, a more subtle process is becoming evident, where small (<10 %) changes to intermediate methylation levels are associated with complex disease phenotypes. This has resulted in two clear methylation paradigms. The latter "subtle change" paradigm is rapidly becoming the epigenetic hallmark of complex disease phenotypes, although we are currently hampered by a lack of data addressing the true biological significance and meaning of these small differences. Our initial expectation of rapidly identifying mechanisms linking environmental exposure to a disease phenotype led to numerous observational/association studies being performed. Although this expectation remains unmet, there is now a growing body of literature on specific genes, suggesting wide ranging transcriptional and translational consequences of such subtle methylation changes. Data from the glucocorticoid receptor (NR3C1) has shown that a complex interplay between DNA methylation, extensive 5'UTR splicing, and microvariability gives rise to the overall level and relative distribution of total and N-terminal protein isoforms generated. Additionally, the presence of multiple AUG translation initiation codons throughout the complete, processed mRNA enables translation variability, hereby enhancing the translational isoforms and the resulting protein isoform diversity, providing a clear link between small changes in DNA methylation and significant changes in protein isoforms and cellular locations. Methylation changes in the NR3C1 CpG island alters the NR3C1 transcription and eventually protein isoforms in the tissues, resulting in subtle but visible physiological variability. This review addresses the current pathophysiological and clinical associations of such characteristically small DNA methylation changes, the ever-growing roles of DNA methylation and the evidence available, particularly from the glucocorticoid receptor of the cascade of events initiated by such subtle methylation changes, as well as addressing the underlying question as to what represents a genuine biologically significant difference in methylation.
Keywords: Association studies; Biomarker; DNA methylation; EWAS; Gene-environment interactions; Transcriptional microvariability.
Figures

 ), transcription factor binding sites (1–25), transcriptional loci (B1–B5), and microvariable transcription start sites (•). Transcription factor binding sites: (
), transcription factor binding sites (1–25), transcriptional loci (B1–B5), and microvariable transcription start sites (•). Transcription factor binding sites: ( ) IRF-1 and IRF-2 (position 1); (
) IRF-1 and IRF-2 (position 1); ( ) glucocorticoid response elements (GRE, positions 2, 3, 8, 11, 14, and 22); (
) glucocorticoid response elements (GRE, positions 2, 3, 8, 11, 14, and 22); ( ) c-Myb, c-Ets1/2 and PU1 (position 4); (
) c-Myb, c-Ets1/2 and PU1 (position 4); ( ) Ying Y and 1 (positions 5, 6, 7, and 25); (■) Sp1 binding sites (positions 9, 10, 12, 13, 16, 19, 20, 21, and 24); (
) Ying Y and 1 (positions 5, 6, 7, and 25); (■) Sp1 binding sites (positions 9, 10, 12, 13, 16, 19, 20, 21, and 24); ( ) Ap-1 (position 15); (
) Ap-1 (position 15); ( ) NGFI-A binding site (position 17); (
) NGFI-A binding site (position 17); ( ) glucocorticoid response factor-1 (GRF-1, position 18); (□) Ap-2 (position 23). b Structure of the GR mRNA with the internal ATG translation initation codons, a western blot demonstrating the different transcriptional isoforms (from [113] with permission), and the frequency of the different protein isoforms with increasing 5′ UTR length (adapted from [111] with permission)
) glucocorticoid response factor-1 (GRF-1, position 18); (□) Ap-2 (position 23). b Structure of the GR mRNA with the internal ATG translation initation codons, a western blot demonstrating the different transcriptional isoforms (from [113] with permission), and the frequency of the different protein isoforms with increasing 5′ UTR length (adapted from [111] with permission)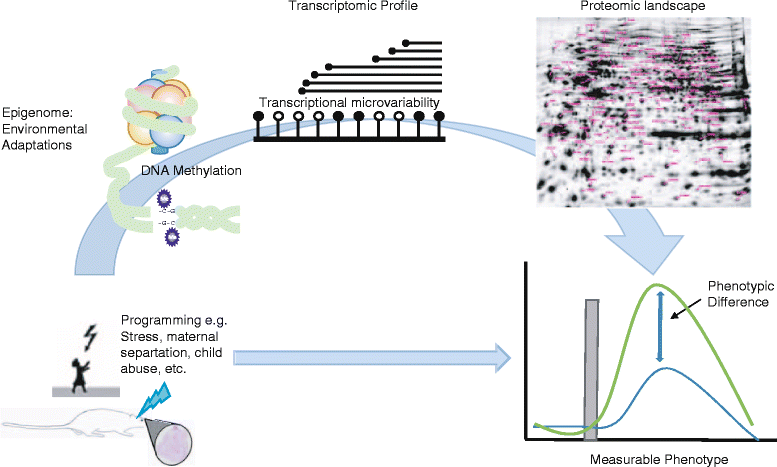
References
-
- Neidhart M. DNA methylation and complex human diseases. 1. Amsterdam: Academic Press; 2016.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

