Temporal dynamics of the developing lung transcriptome in three common inbred strains of laboratory mice reveals multiple stages of postnatal alveolar development
- PMID: 27602285
- PMCID: PMC4991849
- DOI: 10.7717/peerj.2318
Temporal dynamics of the developing lung transcriptome in three common inbred strains of laboratory mice reveals multiple stages of postnatal alveolar development
Abstract
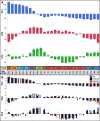
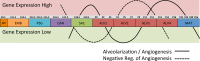
To characterize temporal patterns of transcriptional activity during normal lung development, we generated genome wide gene expression data for 26 pre- and post-natal time points in three common inbred strains of laboratory mice (C57BL/6J, A/J, and C3H/HeJ). Using Principal Component Analysis and least squares regression modeling, we identified both strain-independent and strain-dependent patterns of gene expression. The 4,683 genes contributing to the strain-independent expression patterns were used to define a murine Developing Lung Characteristic Subtranscriptome (mDLCS). Regression modeling of the Principal Components supported the four canonical stages of mammalian embryonic lung development (embryonic, pseudoglandular, canalicular, saccular) defined previously by morphology and histology. For postnatal alveolar development, the regression model was consistent with four stages of alveolarization characterized by episodic transcriptional activity of genes related to pulmonary vascularization. Genes expressed in a strain-dependent manner were enriched for annotations related to neurogenesis, extracellular matrix organization, and Wnt signaling. Finally, a comparison of mouse and human transcriptomics from pre-natal stages of lung development revealed conservation of pathways associated with cell cycle, axon guidance, immune function, and metabolism as well as organism-specific expression of genes associated with extracellular matrix organization and protein modification. The mouse lung development transcriptome data generated for this study serves as a unique reference set to identify genes and pathways essential for normal mammalian lung development and for investigations into the developmental origins of respiratory disease and cancer. The gene expression data are available from the Gene Expression Omnibus (GEO) archive (GSE74243). Temporal expression patterns of mouse genes can be investigated using a study specific web resource (http://lungdevelopment.jax.org).
Keywords: Gene expression; Genomics; Laboratory mice; Lung development; Time series.
Conflict of interest statement
The authors declare there are no competing interests.
Figures



References
-
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Research. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. - DOI - PMC - PubMed
-
- Beller TC, Friend DS, Maekawa A, Lam BK, Austen KF, Kanaoka Y. Cysteinyl leukotriene 1 receptor controls the severity of chronic pulmonary inflammation and fibrosis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3047–3052. doi: 10.1073/pnas.0400235101. - DOI - PMC - PubMed
-
- Bellusci S, Furuta Y, Rush MG, Henderson R, Winnier G, Hogan BL. Involvement of Sonic hedgehog (Shh) in mouse embryonic lung growth and morphogenesis. Development. 1997a;124:53–63. - PubMed
-
- Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL. Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development. 1997b;124:4867–4878. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B. 1995;57:289–300.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

