Extreme Dysbiosis of the Microbiome in Critical Illness
- PMID: 27602409
- PMCID: PMC5007431
- DOI: 10.1128/mSphere.00199-16
Extreme Dysbiosis of the Microbiome in Critical Illness
Abstract
Critical illness is hypothesized to associate with loss of "health-promoting" commensal microbes and overgrowth of pathogenic bacteria (dysbiosis). This dysbiosis is believed to increase susceptibility to nosocomial infections, sepsis, and organ failure. A trial with prospective monitoring of the intensive care unit (ICU) patient microbiome using culture-independent techniques to confirm and characterize this dysbiosis is thus urgently needed. Characterizing ICU patient microbiome changes may provide first steps toward the development of diagnostic and therapeutic interventions using microbiome signatures. To characterize the ICU patient microbiome, we collected fecal, oral, and skin samples from 115 mixed ICU patients across four centers in the United States and Canada. Samples were collected at two time points: within 48 h of ICU admission, and at ICU discharge or on ICU day 10. Sample collection and processing were performed according to Earth Microbiome Project protocols. We applied SourceTracker to assess the source composition of ICU patient samples by using Qiita, including samples from the American Gut Project (AGP), mammalian corpse decomposition samples, childhood (Global Gut study), and house surfaces. Our results demonstrate that critical illness leads to significant and rapid dysbiosis. Many taxons significantly depleted from ICU patients versus AGP healthy controls are key "health-promoting" organisms, and overgrowth of known pathogens was frequent. Source compositions of ICU patient samples are largely uncharacteristic of the expected community type. Between time points and within a patient, the source composition changed dramatically. Our initial results show great promise for microbiome signatures as diagnostic markers and guides to therapeutic interventions in the ICU to repopulate the normal, "health-promoting" microbiome and thereby improve patient outcomes. IMPORTANCE Critical illness may be associated with the loss of normal, "health promoting" bacteria, allowing overgrowth of disease-promoting pathogenic bacteria (dysbiosis), which, in turn, makes patients susceptible to hospital-acquired infections, sepsis, and organ failure. This has significant world health implications, because sepsis is becoming a leading cause of death worldwide, and hospital-acquired infections contribute to significant illness and increased costs. Thus, a trial that monitors the ICU patient microbiome to confirm and characterize this hypothesis is urgently needed. Our study analyzed the microbiomes of 115 critically ill subjects and demonstrated rapid dysbiosis from unexpected environmental sources after ICU admission. These data may provide the first steps toward defining targeted therapies that correct potentially "illness-promoting" dysbiosis with probiotics or with targeted, multimicrobe synthetic "stool pills" that restore a healthy microbiome in the ICU setting to improve patient outcomes.
Keywords: 16S RNA; critical care; fecal organisms; human; microbial source tracking.
Figures
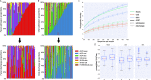
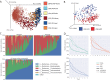
References
-
- Gevers D, Kugathasan S, Denson L, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song S, Yassour M, Morgan X, Kostic A, Luo C, González A, McDonald D, Haberman Y, Walters T, Baker S, Rosh J, Stephens M, Heyman M, Markowitz J, Baldassano R, Griffiths A, Sylvester F, Mack D, Kim S, Crandall W, Hyams J, Huttenhower C, Knight R, Xavier RJ. 2014. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15:382–392. doi: 10.1016/j.chom.2014.02.005. - DOI - PMC - PubMed
-
- Weingarden A, González A, Vázquez-Baeza Y, Weiss S, Humphry G, Berg-Lyons D, Knights D, Unno T, Bobr A, Kang J, Khoruts A, Knight R, Sadowsky MJ. 2015. Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome 3:10. doi: 10.1186/s40168-015-0070-0. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

