Quantification of nucleic acid quality in postmortem tissues from a cancer research autopsy program
- PMID: 27602498
- PMCID: PMC5341846
- DOI: 10.18632/oncotarget.11836
Quantification of nucleic acid quality in postmortem tissues from a cancer research autopsy program
Abstract
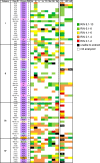
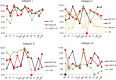
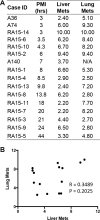
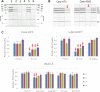
The last decade has seen a marked rise in the use of cancer tissues obtained from research autopsies. Such resources have been invaluable for studying cancer evolution or the mechanisms of therapeutic resistance to targeted therapies. Degradation of biomolecules is a potential challenge to usage of cancer tissues obtained in the post-mortem setting and remains incompletely studied. We analysed the nucleic acid quality in 371 different frozen tissue samples collected from 80 patients who underwent a research autopsy, including eight normal tissue types, primary and metastatic tumors. Our results indicate that RNA integrity number (RIN) of normal tissues decline with the elongation of post-mortem interval (PMI) in a tissue-type specific manner. Unlike normal tissues, the RNA quality of cancer tissues is highly variable with respect to post-mortem interval. The kinetics of DNA damage also has tissue type-specific features. Moreover, while DNA degradation is an indicator of low RNA quality, the converse is not true. Finally, we show that despite RIN values as low as 5.0, robust data can be obtained by RNA sequencing that reliably discriminates expression signatures.
Keywords: RNA; RNA sequencing; autopsy; metastasis; post-mortem.
Conflict of interest statement
The authors have declared no potential conflicts of interest.
Figures




References
-
- Clark MJ. Autopsy. Lancet. 2005;366:1767. - PubMed
-
- Scatliff JH, Morris PJ. From Roentgen to magnetic resonance imaging: the history of medical imaging. N C Med J. 2014;75:111–113. - PubMed
-
- Burke MD. Laboratory medicine in the 21st Century. Am J Clin Pathol. 2000;114:841–846. - PubMed
-
- Samarasekera N, Al-Shahi Salman R, Huitinga I, Klioueva N, McLean CA, Kretzschmar H, Smith C, Ironside JW. Brain banking for neurological disorders. Lancet Neurol. 2013;12:1096–1105. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

