Genomic Characterization of the Novel Aeromonas hydrophila Phage Ahp1 Suggests the Derivation of a New Subgroup from phiKMV-Like Family
- PMID: 27603936
- PMCID: PMC5014404
- DOI: 10.1371/journal.pone.0162060
Genomic Characterization of the Novel Aeromonas hydrophila Phage Ahp1 Suggests the Derivation of a New Subgroup from phiKMV-Like Family
Abstract
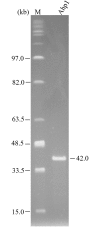
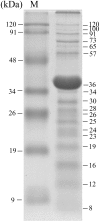
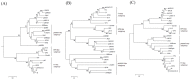
Aeromonas hydrophila is an opportunistic pathogenic bacterium causing diseases in human and fish. The emergence of multidrug-resistant A. hydrophila isolates has been increasing in recent years. In this study, we have isolated a novel virulent podophage of A. hydrophila, designated as Ahp1, from waste water. Ahp1 has a rapid adsorption (96% adsorbed in 2 min), a latent period of 15 min, and a burst size of 112 PFU per infected cell. At least eighteen Ahp1 virion proteins were visualized in SDS-polyacrylamide gel electrophoresis, with a 36-kDa protein being the predicted major capsid protein. Genome analysis of Ahp1 revealed a linear doubled-stranded DNA genome of 42,167 bp with a G + C content of 58.8%. The genome encodes 46 putative open reading frames, 5 putative phage promoters, and 3 transcriptional terminators. Based on high degrees of similarity in overall genome organization and among most of the corresponding ORFs, as well as phylogenetic relatedness among their DNAP, RNAP and major capsid proteins, we propose a new subgroup, designated Ahp1-like subgroup. This subgroup contains Ahp1 and members previously belonging to phiKMV-like subgroup, phiAS7, phi80-18, GAP227, phiR8-01, and ISAO8. Since Ahp1 has a narrow host range, for effective phage therapy, different phages are needed for preparation of cocktails that are capable of killing the heterogeneous A. hydrophila strains.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Molecular Characterization of Ahp2, a Lytic Bacteriophage of Aeromonas hydrophila.Viruses. 2021 Mar 14;13(3):477. doi: 10.3390/v13030477. Viruses. 2021. PMID: 33799428 Free PMC article.
-
Characterization and genomic analyses of Aeromonas hydrophila phages AhSzq-1 and AhSzw-1, isolates representing new species within the T5virus genus.Arch Virol. 2018 Jul;163(7):1985-1988. doi: 10.1007/s00705-018-3805-y. Epub 2018 Mar 19. Arch Virol. 2018. PMID: 29556775
-
Isolation, characterization and genomic analysis of vB-AhyM-AP1, a lytic bacteriophage infecting Aeromonas hydrophila.J Appl Microbiol. 2021 Aug;131(2):695-705. doi: 10.1111/jam.14997. Epub 2021 Jan 23. J Appl Microbiol. 2021. PMID: 33420733
-
A virulent phage infecting Lactococcus garvieae, with homology to Lactococcus lactis phages.Appl Environ Microbiol. 2015 Dec;81(24):8358-65. doi: 10.1128/AEM.02603-15. Epub 2015 Sep 25. Appl Environ Microbiol. 2015. PMID: 26407890 Free PMC article.
-
Genomic characterization of a novel virulent phage infecting the Aeromonas hydrophila isolated from rainbow trout (Oncorhynchus mykiss).Virus Res. 2019 Nov;273:197764. doi: 10.1016/j.virusres.2019.197764. Epub 2019 Sep 21. Virus Res. 2019. PMID: 31550486
Cited by
-
Characterization and diversity of phages infecting Aeromonas salmonicida subsp. salmonicida.Sci Rep. 2017 Aug 1;7(1):7054. doi: 10.1038/s41598-017-07401-7. Sci Rep. 2017. PMID: 28765570 Free PMC article.
-
Characterization and Genome Analyses of the Novel Phages P2 and vB_AhydM-H1 Targeting Aeromonas hydrophila.Phage (New Rochelle). 2024 Sep 16;5(3):162-172. doi: 10.1089/phage.2024.0014. eCollection 2024 Sep. Phage (New Rochelle). 2024. PMID: 39372357
-
Nine Novel Phages from a Plateau Lake in Southwest China: Insights into Aeromonas Phage Diversity.Viruses. 2019 Jul 5;11(7):615. doi: 10.3390/v11070615. Viruses. 2019. PMID: 31284428 Free PMC article.
-
A Novel Aeromonas popoffii Phage AerP_220 Proposed to Be a Member of a New Tolavirus Genus in the Autographiviridae Family.Viruses. 2022 Dec 7;14(12):2733. doi: 10.3390/v14122733. Viruses. 2022. PMID: 36560737 Free PMC article.
-
Characterization of Novel Bacteriophage AhyVDH1 and Its Lytic Activity Against Aeromonas hydrophila.Curr Microbiol. 2021 Jan;78(1):329-337. doi: 10.1007/s00284-020-02279-7. Epub 2020 Nov 11. Curr Microbiol. 2021. PMID: 33175194
References
-
- Alavandi SV, Subashini MS, Ananthan S. Occurrence of haemolytic & cytotoxic Aeromonas species in domestic water supplies in Chennai. The Indian journal of medical research. 1999;110:50–5. . - PubMed
-
- Chauret C, Volk C, Creason R, Jarosh J, Robinson J, Warnes C. Detection of Aeromonas hydrophila in a drinking-water distribution system: a field and pilot study. Canadian journal of microbiology. 2001;47(8):782–6. . - PubMed
-
- Pablos M, Remacha MA, Rodriguez-Calleja JM, Santos JA, Otero A, Garcia-Lopez ML. Identity, virulence genes, and clonal relatedness of Aeromonas isolates from patients with diarrhea and drinking water. European journal of clinical microbiology & infectious diseases: official publication of the European Society of Clinical Microbiology. 2010;29(9):1163–72. 10.1007/s10096-010-0982-3 . - DOI - PubMed
-
- Danaher PJ, Mueller WP. Aeromonas hydrophila septic arthritis. Military medicine. 2011;176(12):1444–6. . - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

