NSD2 contributes to oncogenic RAS-driven transcription in lung cancer cells through long-range epigenetic activation
- PMID: 27604143
- PMCID: PMC5015087
- DOI: 10.1038/srep32952
NSD2 contributes to oncogenic RAS-driven transcription in lung cancer cells through long-range epigenetic activation
Abstract
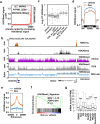
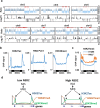
The histone methyltransferase NSD2/WHSC1/MMSET is overexpressed in a number of solid tumors but its contribution to the biology of these tumors is not well understood. Here, we describe that NSD2 contributes to the proliferation of a subset of lung cancer cell lines by supporting oncogenic RAS transcriptional responses. NSD2 knock down combined with MEK or BRD4 inhibitors causes co-operative inhibitory responses on cell growth. However, while MEK and BRD4 inhibitors converge in the downregulation of genes associated with cancer-acquired super-enhancers, NSD2 inhibition affects the expression of clusters of genes embedded in megabase-scale regions marked with H3K36me2 and that contribute to the RAS transcription program. Thus, combinatorial therapies using MEK or BRD4 inhibitors together with NSD2 inhibition are likely to be needed to ensure a more comprehensive inhibition of oncogenic RAS-driven transcription programs in lung cancers with NSD2 overexpression.
Conflict of interest statement
This work was supported by Eli Lilly and Company. B.H., H.B.B. and R.M.C. are employees of Eli Lilly and Company.
Figures
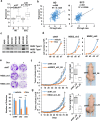
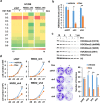
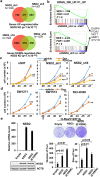
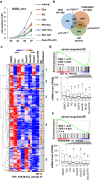
References
-
- Morishita M. & Di Luccio E. Cancers and the NSD family of histone lysine methyltransferases. Biochim. Biophys. Acta - Rev. Cancer 1816, 158–163 (2011). - PubMed
-
- Stec I. et al. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum. Mol. Genet. 7, 1071–82 (1998). - PubMed
-
- Chesi M. et al. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood 92, 3025–34 (1998). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

