Mapping the complete glycoproteome of virion-derived HIV-1 gp120 provides insights into broadly neutralizing antibody binding
- PMID: 27604319
- PMCID: PMC5015092
- DOI: 10.1038/srep32956
Mapping the complete glycoproteome of virion-derived HIV-1 gp120 provides insights into broadly neutralizing antibody binding
Abstract
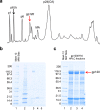
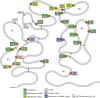
The surface envelope glycoprotein (SU) of Human immunodeficiency virus type 1 (HIV-1), gp120(SU) plays an essential role in virus binding to target CD4+ T-cells and is a major vaccine target. Gp120 has remarkably high levels of N-linked glycosylation and there is considerable evidence that this "glycan shield" can help protect the virus from antibody-mediated neutralization. In recent years, however, it has become clear that gp120 glycosylation can also be included in the targets of recognition by some of the most potent broadly neutralizing antibodies. Knowing the site-specific glycosylation of gp120 can facilitate the rational design of glycopeptide antigens for HIV vaccine development. While most prior studies have focused on glycan analysis of recombinant forms of gp120, here we report the first systematic glycosylation site analysis of gp120 derived from virions produced by infected T lymphoid cells and show that a single site is exclusively substituted with complex glycans. These results should help guide the design of vaccine immunogens.
Figures


References
-
- Zhu X., Borchers C., Bienstock R. J. & Tomer K. B. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry 39, 11194–11204 (2000). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

