High-Sequence Diversity and Rapid Virus Turnover Contribute to Higher Rates of Coreceptor Switching in Treatment-Experienced Subjects with HIV-1 Viremia
- PMID: 27604829
- PMCID: PMC5333564
- DOI: 10.1089/AID.2016.0153
High-Sequence Diversity and Rapid Virus Turnover Contribute to Higher Rates of Coreceptor Switching in Treatment-Experienced Subjects with HIV-1 Viremia
Abstract
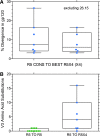
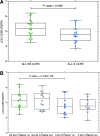
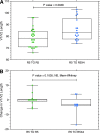
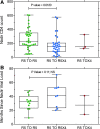
Coreceptor switching from CCR5 to CXCR4 is common during chronic HIV-1 infection, but is even more common in individuals who have failed antiretroviral therapy (ART). Prior studies have suggested rapid mutation and/or recombination of HIV-1 envelope (env) genes during coreceptor switching. We compared the functional and genotypic changes in env of viruses from viremic subjects who had failed ART just before and after coreceptor switching and compared those to viruses from matched subjects without coreceptor switching. Analysis of multiple unique functional env clones from each subject revealed extensive diversity at both sample time points and rapid diversification of sequences during the 4-month interval in viruses from both 9 subjects with coreceptor switching and 15 control subjects. Only two subjects had envs with evidence of recombination. Three findings distinguished env clones from subjects with coreceptor switching from controls: (1) lower entry efficiency via CCR5; (2) longer V1/V2 regions; and (3), lower nadir CD4 T cell counts during prior years of infection. Most of these subjects harbored virus with lower replicative capacity associated with protease (PR) and/or reverse transcriptase inhibitor resistance mutations, and the extensive diversification tended to lead either to improved entry efficiency via CCR5 or the gain of entry function via CXCR4. These results suggest that R5X4 or X4 variants emerge from a diverse, low-fitness landscape shaped by chronic infection, multiple ART resistance mutations, the availability of target cells, and reduced entry efficiency via CCR5.
Keywords: HIV-1 coreceptor switching; sequence diversity; sequence evolution.
Conflict of interest statement
J.D.R. and E.D.A. are employees of Monogram Biosciences, Laboratory Corporation of America Holdings (LH), and shareholders of LH. The other authors report no conflicts of interest exist.
Figures

Similar articles
-
Linkages between HIV-1 specificity for CCR5 or CXCR4 and in vitro usage of alternative coreceptors during progressive HIV-1 subtype C infection.Retrovirology. 2013 Sep 16;10:98. doi: 10.1186/1742-4690-10-98. Retrovirology. 2013. PMID: 24041034 Free PMC article.
-
Differences in molecular evolution between switch (R5 to R5X4/X4-tropic) and non-switch (R5-tropic only) HIV-1 populations during infection.Infect Genet Evol. 2010 Apr;10(3):356-64. doi: 10.1016/j.meegid.2009.05.003. Epub 2009 May 14. Infect Genet Evol. 2010. PMID: 19446658
-
Deep Sequencing of the HIV-1 env Gene Reveals Discrete X4 Lineages and Linkage Disequilibrium between X4 and R5 Viruses in the V1/V2 and V3 Variable Regions.J Virol. 2016 Jul 27;90(16):7142-58. doi: 10.1128/JVI.00441-16. Print 2016 Aug 15. J Virol. 2016. PMID: 27226378 Free PMC article.
-
How HIV changes its tropism: evolution and adaptation?Curr Opin HIV AIDS. 2009 Mar;4(2):125-30. doi: 10.1097/COH.0b013e3283223d61. Curr Opin HIV AIDS. 2009. PMID: 19339951 Free PMC article. Review.
-
Genotypic coreceptor analysis.Eur J Med Res. 2007 Oct 15;12(9):453-62. Eur J Med Res. 2007. PMID: 17933727 Review.
Cited by
-
Korean Red Ginseng slows coreceptor switch in HIV-1 infected patients.J Ginseng Res. 2023 Jan;47(1):117-122. doi: 10.1016/j.jgr.2022.06.003. Epub 2022 Jul 6. J Ginseng Res. 2023. PMID: 36644395 Free PMC article.
-
HIV tropism switch in archived DNA of HIV-HCV subjects successfully treated with direct-acting antivirals for HCV infection.Sci Rep. 2021 Apr 29;11(1):9274. doi: 10.1038/s41598-021-88811-6. Sci Rep. 2021. PMID: 33927306 Free PMC article.
References
-
- Berger EA, Murphy PM, Farber JM: Chemokine receptors as HIV-1 coreceptors: Roles in viral entry, tropism, and disease. Annu Rev Immunol 1999;17:657–700 - PubMed
-
- Cocchi F, DeVico AL, Garzino-Demo A, Cara A, Gallo RC, Lusso P: The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med 1996;2:1244–1247 - PubMed
-
- Hwang SS, Boyle TJ, Lyerly HK, Cullen BR: Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science 1991;253:71–74 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

