Alu repeats as transcriptional regulatory platforms in macrophage responses to M. tuberculosis infection
- PMID: 27604870
- PMCID: PMC5159539
- DOI: 10.1093/nar/gkw782
Alu repeats as transcriptional regulatory platforms in macrophage responses to M. tuberculosis infection
Abstract
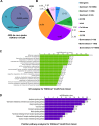
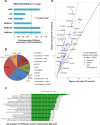
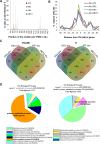
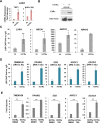
To understand the epigenetic regulation of transcriptional response of macrophages during early-stage M. tuberculosis (Mtb) infection, we performed ChIPseq analysis of H3K4 monomethylation (H3K4me1), a marker of poised or active enhancers. De novo H3K4me1 peaks in infected cells were associated with genes implicated in host defenses and apoptosis. Our analysis revealed that 40% of de novo regions contained human/primate-specific Alu transposable elements, enriched in the AluJ and S subtypes. These contained several transcription factor binding sites, including those for members of the MEF2 and ATF families, and LXR and RAR nuclear receptors, all of which have been implicated in macrophage differentiation, survival, and responses to stress and infection. Combining bioinformatics, molecular genetics, and biochemical approaches, we linked genes adjacent to H3K4me1-associated Alu repeats to macrophage metabolic responses against Mtb infection. In particular, we show that LXRα signaling, which reduced Mtb viability 18-fold by altering cholesterol metabolism and enhancing macrophage apoptosis, can be initiated at response elements present in Alu repeats. These studies decipher the mechanism of early macrophage transcriptional responses to Mtb, highlighting the role of Alu element transposition in shaping human transcription programs during innate immunity.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
The Transcriptional Foundations of Sp110-mediated Macrophage (RAW264.7) Resistance to Mycobacterium tuberculosis H37Ra.Sci Rep. 2016 Feb 25;6:22041. doi: 10.1038/srep22041. Sci Rep. 2016. PMID: 26912204 Free PMC article.
-
Trans-species communication in the Mycobacterium tuberculosis-infected macrophage.Immunol Rev. 2015 Mar;264(1):233-48. doi: 10.1111/imr.12254. Immunol Rev. 2015. PMID: 25703563 Free PMC article. Review.
-
Immunoevasion and immunosuppression of the macrophage by Mycobacterium tuberculosis.Immunol Rev. 2015 Mar;264(1):220-32. doi: 10.1111/imr.12268. Immunol Rev. 2015. PMID: 25703562 Review.
-
Transcriptionally induced enhancers in the macrophage immune response to Mycobacterium tuberculosis infection.BMC Genomics. 2019 Jan 22;20(1):71. doi: 10.1186/s12864-019-5450-6. BMC Genomics. 2019. PMID: 30669987 Free PMC article.
-
Engagement of the Aryl Hydrocarbon Receptor in Mycobacterium tuberculosis-Infected Macrophages Has Pleiotropic Effects on Innate Immune Signaling.J Immunol. 2015 Nov 1;195(9):4479-91. doi: 10.4049/jimmunol.1501141. Epub 2015 Sep 28. J Immunol. 2015. PMID: 26416282
Cited by
-
Many Facets of Dynamic Plasticity in Plants.Cold Spring Harb Perspect Biol. 2019 Oct 1;11(10):a034629. doi: 10.1101/cshperspect.a034629. Cold Spring Harb Perspect Biol. 2019. PMID: 31138545 Free PMC article. Review.
-
Macrophage nuclear receptors: Emerging key players in infectious diseases.PLoS Pathog. 2019 Mar 21;15(3):e1007585. doi: 10.1371/journal.ppat.1007585. eCollection 2019 Mar. PLoS Pathog. 2019. PMID: 30897154 Free PMC article. Review.
-
Tissue and cancer-specific expression of DIEXF is epigenetically mediated by an Alu repeat.Epigenetics. 2020 Jun-Jul;15(6-7):765-779. doi: 10.1080/15592294.2020.1722398. Epub 2020 Feb 11. Epigenetics. 2020. PMID: 32041475 Free PMC article.
-
A Multilayered Control of the Human Survival Motor Neuron Gene Expression by Alu Elements.Front Microbiol. 2017 Nov 15;8:2252. doi: 10.3389/fmicb.2017.02252. eCollection 2017. Front Microbiol. 2017. PMID: 29187847 Free PMC article. Review.
-
A new human embryonic cell type associated with activity of young transposable elements allows definition of the inner cell mass.PLoS Biol. 2023 Jun 20;21(6):e3002162. doi: 10.1371/journal.pbio.3002162. eCollection 2023 Jun. PLoS Biol. 2023. PMID: 37339119 Free PMC article.
References
-
- Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. - PubMed
-
- Hedges D.J., Batzer M.A. From the margins of the genome: mobile elements shape primate evolution. BioEssays News Rev. Mol. Cell. Dev. Biol. 2005;27:785–794. - PubMed
-
- Canté-Barrett K., Pieters R., Meijerink J.P.P. Myocyte enhancer factor 2C in hematopoiesis and leukemia. Oncogene. 2014;33:403–410. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

