Predicting the Stability of Homologous Gene Duplications in a Plant RNA Virus
- PMID: 27604880
- PMCID: PMC5633665
- DOI: 10.1093/gbe/evw219
Predicting the Stability of Homologous Gene Duplications in a Plant RNA Virus
Abstract
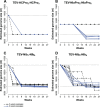
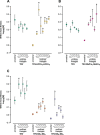
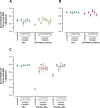
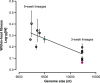
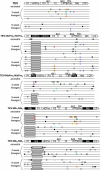
One of the striking features of many eukaryotes is the apparent amount of redundancy in coding and non-coding elements of their genomes. Despite the possible evolutionary advantages, there are fewer examples of redundant sequences in viral genomes, particularly those with RNA genomes. The factors constraining the maintenance of redundant sequences in present-day RNA virus genomes are not well known. Here, we use Tobacco etch virus, a plant RNA virus, to investigate the stability of genetically redundant sequences by generating viruses with potentially beneficial gene duplications. Subsequently, we tested the viability of these viruses and performed experimental evolution. We found that all gene duplication events resulted in a loss of viability or in a significant reduction in viral fitness. Moreover, upon analyzing the genomes of the evolved viruses, we always observed the deletion of the duplicated gene copy and maintenance of the ancestral copy. Interestingly, there were clear differences in the deletion dynamics of the duplicated gene associated with the passage duration and the size and position of the duplicated copy. Based on the experimental data, we developed a mathematical model to characterize the stability of genetically redundant sequences, and showed that fitness effects are not enough to predict genomic stability. A context-dependent recombination rate is also required, with the context being the duplicated gene and its position. Our results therefore demonstrate experimentally the deleterious nature of gene duplications in RNA viruses. Beside previously described constraints on genome size, we identified additional factors that reduce the likelihood of the maintenance of duplicated genes.
Keywords: experimental evolution; gene duplication; genome stability; virus evolution.
© The Author 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
-
- Andersson DI, Hughes D. 2009. Gene amplification and adaptive evolution in bacteria. Annu. Rev. Genet. 43:167–195. - PubMed
-
- Atreya CD, Atreya PL, Thornbury DW, Pirone TP. 1992. Site-directed mutations in the potyvirus HC-PRO gene affect helper component activity, virus accumulation, and symptom expression in infected tobacco plants. Virology 191:106–111. - PubMed
-
- Bedoya LC, Daròs J-A. 2010. Stability of Tobacco etch virus infectious clones in plasmid vectors. Virus Res. 149:234–240. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

