TGF-β-induced stromal CYR61 promotes resistance to gemcitabine in pancreatic ductal adenocarcinoma through downregulation of the nucleoside transporters hENT1 and hCNT3
- PMID: 27604902
- PMCID: PMC5091039
- DOI: 10.1093/carcin/bgw093
TGF-β-induced stromal CYR61 promotes resistance to gemcitabine in pancreatic ductal adenocarcinoma through downregulation of the nucleoside transporters hENT1 and hCNT3
Abstract
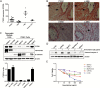
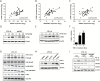
Pancreatic ductal adenocarcinoma (PDAC) is a lethal cancer in part due to inherent resistance to chemotherapy, including the first-line drug gemcitabine. Although low expression of the nucleoside transporters hENT1 and hCNT3 that mediate cellular uptake of gemcitabine has been linked to gemcitabine resistance, the mechanisms regulating their expression in the PDAC tumor microenvironment are largely unknown. Here, we report that the matricellular protein cysteine-rich angiogenic inducer 61 (CYR61) negatively regulates the nucleoside transporters hENT1 and hCNT3. CRISPR/Cas9-mediated knockout of CYR61 increased expression of hENT1 and hCNT3, increased cellular uptake of gemcitabine and sensitized PDAC cells to gemcitabine-induced apoptosis. In PDAC patient samples, expression of hENT1 and hCNT3 negatively correlates with expression of CYR61 . We demonstrate that stromal pancreatic stellate cells (PSCs) are a source of CYR61 within the PDAC tumor microenvironment. Transforming growth factor-β (TGF-β) induces the expression of CYR61 in PSCs through canonical TGF-β-ALK5-Smad2/3 signaling. Activation of TGF-β signaling or expression of CYR61 in PSCs promotes resistance to gemcitabine in PDAC cells in an in vitro co-culture assay. Our results identify CYR61 as a TGF-β-induced stromal-derived factor that regulates gemcitabine sensitivity in PDAC and suggest that targeting CYR61 may improve chemotherapy response in PDAC patients.
© The Author 2016. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures




References
-
- Siegel R., et al. (2014) Cancer statistics, 2014. CA: A Cancer J. Clin., 64, 9–29. - PubMed
-
- Rahib L., et al. (2014) Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res., 74, 2913–2921. - PubMed
-
- de Sousa Cavalcante L., et al. (2014) Gemcitabine: metabolism and molecular mechanisms of action, sensitivity and chemoresistance in pancreatic cancer. Eur. J. Pharmacol., 741, 8–16. - PubMed
-
- Burris H.A., 3rd, et al. (1997) Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J. Clin. Oncol., 15, 2403–2413. - PubMed
-
- Garrido-Laguna I., et al. (2015) Pancreatic cancer: from state-of-the-art treatments to promising novel therapies. Nat. Rev. Clin. Oncol., 12, 319–334. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

