Phenotypic MicroRNA Microarrays
- PMID: 27605181
- PMCID: PMC5003479
- DOI: 10.3390/microarrays2020063
Phenotypic MicroRNA Microarrays
Abstract
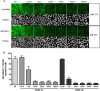
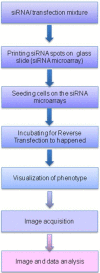
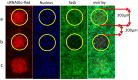
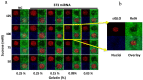
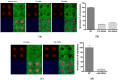
Microarray technology has become a very popular approach in cases where multiple experiments need to be conducted repeatedly or done with a variety of samples. In our lab, we are applying our high density spots microarray approach to microscopy visualization of the effects of transiently introduced siRNA or cDNA on cellular morphology or phenotype. In this publication, we are discussing the possibility of using this micro-scale high throughput process to study the role of microRNAs in the biology of selected cellular models. After reverse-transfection of microRNAs and siRNA, the cellular phenotype generated by microRNAs regulated NF-κB expression comparably to the siRNA. The ability to print microRNA molecules for reverse transfection into cells is opening up the wide horizon for the phenotypic high content screening of microRNA libraries using cellular disease models.
Keywords: microRNA; phenotypic screen; siRNA.
Figures




References
-
- Elbashir S.M., Harborth J., Lendeckel W., Yalch A., Weber K., Tuschl T. Duplex of 21-nucleotide RNAs mediates RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

