Copy Number Studies in Noisy Samples
- PMID: 27605193
- PMCID: PMC5003442
- DOI: 10.3390/microarrays2040284
Copy Number Studies in Noisy Samples
Abstract
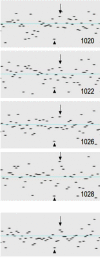
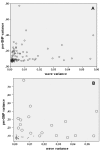
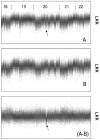
System noise was analyzed in 77 Affymetrix 6.0 samples from a previous clinical study of copy number variation (CNV). Twenty-three samples were classified as eligible for CNV detection, 29 samples as ineligible and 25 were classified as being of intermediate quality. New software ("noise-free-cnv") was developed to visualize the data and reduce system noise. Fresh DNA preparations were more likely to yield eligible samples (p < 0.001). Eligible samples had higher rates of successfully genotyped SNPs (p < 0.001) and lower variance of signal intensities (p < 0.001), yielded fewer CNV findings after Birdview analysis (p < 0.001), and showed a tendency to yield fewer PennCNV calls (p = 0.053). The noise-free-cnv software visualized trend patterns of noise in the signal intensities across the ordered SNPs, including a wave pattern of noise, being co-linear with the banding pattern of metaphase chromosomes, as well as system deviations of individual probe sets (per-SNP noise). Wave noise and per-SNP noise occurred independently and could be separately removed from the samples. We recommend a two-step procedure of CNV validation, including noise reduction and visual inspection of all CNV calls, prior to molecular validation of a selected number of putative CNVs.
Keywords: copy number variation (CNV); noise reduction; noise-free-cnv software; per-SNP noise; validation of CNV findings; variance; wave noise.
Figures






References
-
- Fakhro K.A., Choi M., Ware S.M., Belmont J.W., Towbin J.A., Lifton R.P., Khokha M.K., Brueckner M. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proc. Natl. Acad. Sci. USA. 2011;108:2915–2920. doi: 10.1073/pnas.1019645108. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

