p38 (Mapk14/11) occupies a regulatory node governing entry into primitive endoderm differentiation during preimplantation mouse embryo development
- PMID: 27605380
- PMCID: PMC5043583
- DOI: 10.1098/rsob.160190
p38 (Mapk14/11) occupies a regulatory node governing entry into primitive endoderm differentiation during preimplantation mouse embryo development
Abstract
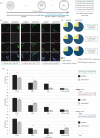
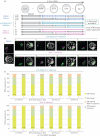
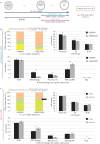
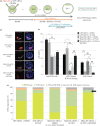
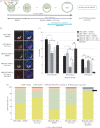
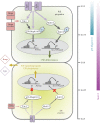
During mouse preimplantation embryo development, the classically described second cell-fate decision involves the specification and segregation, in blastocyst inner cell mass (ICM), of primitive endoderm (PrE) from pluripotent epiblast (EPI). The active role of fibroblast growth factor (Fgf) signalling during PrE differentiation, particularly in the context of Erk1/2 pathway activation, is well described. However, we report that p38 family mitogen-activated protein kinases (namely p38α/Mapk14 and p38β/Mapk11; referred to as p38-Mapk14/11) also participate in PrE formation. Specifically, functional p38-Mapk14/11 are required, during early-blastocyst maturation, to assist uncommitted ICM cells, expressing both EPI and earlier PrE markers, to fully commit to PrE differentiation. Moreover, functional activation of p38-Mapk14/11 is, as reported for Erk1/2, under the control of Fgf-receptor signalling, plus active Tak1 kinase (involved in non-canonical bone morphogenetic protein (Bmp)-receptor-mediated PrE differentiation). However, we demonstrate that the critical window of p38-Mapk14/11 activation precedes the E3.75 timepoint (defined by the initiation of the classical 'salt and pepper' expression pattern of mutually exclusive EPI and PrE markers), whereas appropriate lineage maturation is still achievable when Erk1/2 activity (via Mek1/2 inhibition) is limited to a period after E3.75. We propose that active p38-Mapk14/11 act as enablers, and Erk1/2 as drivers, of PrE differentiation during ICM lineage specification and segregation.
Keywords: cell signalling; cell-fate; mitogen-activated protein kinase; p38α/p38β Mapk14/Mapk11; preimplantation mouse embryo; primitive endoderm.
© 2016 The Authors.
Figures
References
-
- Rossant J, Tam PP. 2009. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development 136, 701–713. (doi:10.1242/dev.017178) - DOI - PubMed
-
- Schrode N, Xenopoulos P, Piliszek A, Frankenberg S, Plusa B, Hadjantonakis AK. 2013. Anatomy of a blastocyst: cell behaviors driving cell fate choice and morphogenesis in the early mouse embryo. Genesis 51, 219–233. (doi:10.1002/dvg.22368) - DOI - PMC - PubMed
-
- Zernicka-Goetz M, Morris SA, Bruce AW. 2009. Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat. Rev. Genet. 10, 467–477. (doi:10.1038/nrg2564) - DOI - PubMed
-
- Sasaki H. 2015. Position- and polarity-dependent Hippo signaling regulates cell fates in preimplantation mouse embryos. Semin. Cell Dev. Biol. 48, 80–87. (doi:10.1016/j.semcdb.2015.05.003) - DOI - PubMed
-
- Nishioka N, Yamamoto S, Kiyonari H, Sato H, Sawada A, Ota M, Nakao K, Sasaki H. 2008. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech. Dev. 125, 270–283. (doi:10.1016/j.mod.2007.11.002) - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
