Structural insights into Parkin substrate lysine targeting from minimal Miro substrates
- PMID: 27605430
- PMCID: PMC5015425
- DOI: 10.1038/srep33019
Structural insights into Parkin substrate lysine targeting from minimal Miro substrates
Abstract
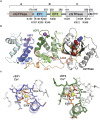
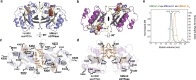
Hereditary Parkinson's disease is commonly caused by mutations in the protein kinase PINK1 or the E3 ubiquitin ligase Parkin, which function together to eliminate damaged mitochondria. PINK1 phosphorylates both Parkin and ubiquitin to stimulate ubiquitination of dozens of proteins on the surface of the outer mitochondrial membrane. However, the mechanisms by which Parkin recognizes specific proteins for modification remain largely unexplored. Here, we show that the C-terminal GTPase (cGTPase) of the Parkin primary substrate human Miro is necessary and sufficient for efficient ubiquitination. We present several new X-ray crystal structures of both human Miro1 and Miro2 that reveal substrate recognition and ubiquitin transfer to be specific to particular protein domains and lysine residues. We also provide evidence that Parkin substrate recognition is functionally separate from substrate modification. Finally, we show that prioritization for modification of a specific lysine sidechain of the cGTPase (K572) within human Miro1 is dependent on both its location and chemical microenvironment. Activation of Parkin by phosphorylation or by binding of pUb is required for prioritization of K572 for modification, suggesting that Parkin activation and acquisition of substrate specificity are coupled.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

