Sublethal salinity stress contributes to habitat limitation in an endangered estuarine fish
- PMID: 27606005
- PMCID: PMC4999527
- DOI: 10.1111/eva.12385
Sublethal salinity stress contributes to habitat limitation in an endangered estuarine fish
Abstract
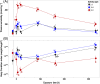
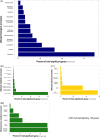
As global change alters multiple environmental conditions, predicting species' responses can be challenging without understanding how each environmental factor influences organismal performance. Approaches quantifying mechanistic relationships can greatly complement correlative field data, strengthening our abilities to forecast global change impacts. Substantial salinity increases are projected in the San Francisco Estuary, California, due to anthropogenic water diversion and climatic changes, where the critically endangered delta smelt (Hypomesus transpacificus) largely occurs in a low-salinity zone (LSZ), despite their ability to tolerate a much broader salinity range. In this study, we combined molecular and organismal measures to quantify the physiological mechanisms and sublethal responses involved in coping with salinity changes. Delta smelt utilize a suite of conserved molecular mechanisms to rapidly adjust their osmoregulatory physiology in response to salinity changes in estuarine environments. However, these responses can be energetically expensive, and delta smelt body condition was reduced at high salinities. Thus, acclimating to salinities outside the LSZ could impose energetic costs that constrain delta smelt's ability to exploit these habitats. By integrating data across biological levels, we provide key insight into the mechanistic relationships contributing to phenotypic plasticity and distribution limitations and advance the understanding of the molecular osmoregulatory responses in nonmodel estuarine fishes.
Keywords: Hypomesus transpacificus; anadromous fish; climate change; delta smelt; environmental stress; osmoregulation; transcriptome.
Figures




Similar articles
-
Turbidity and salinity affect feeding performance and physiological stress in the endangered delta smelt.Integr Comp Biol. 2013 Oct;53(4):620-34. doi: 10.1093/icb/ict082. Epub 2013 Aug 5. Integr Comp Biol. 2013. PMID: 23922273
-
Ontogeny influences sensitivity to climate change stressors in an endangered fish.Conserv Physiol. 2014 Mar 10;2(1):cou008. doi: 10.1093/conphys/cou008. eCollection 2014. Conserv Physiol. 2014. PMID: 27293629 Free PMC article.
-
Sensitivities of an endemic, endangered California smelt and two non-native fishes to serial increases in temperature and salinity: implications for shifting community structure with climate change.Conserv Physiol. 2019 Feb 18;7(1):coy076. doi: 10.1093/conphys/coy076. eCollection 2019. Conserv Physiol. 2019. PMID: 30842886 Free PMC article.
-
The physiology of hyper-salinity tolerance in teleost fish: a review.J Comp Physiol B. 2012 Apr;182(3):321-9. doi: 10.1007/s00360-011-0624-9. Epub 2011 Oct 28. J Comp Physiol B. 2012. PMID: 22033744 Review.
-
Physiological mechanisms used by fish to cope with salinity stress.J Exp Biol. 2015 Jun;218(Pt 12):1907-14. doi: 10.1242/jeb.118695. J Exp Biol. 2015. PMID: 26085667 Review.
Cited by
-
Contaminant exposure effects in a changing climate: how multiple stressors can multiply exposure effects in the amphipod Hyalella azteca.Ecotoxicology. 2018 Sep;27(7):845-859. doi: 10.1007/s10646-018-1912-x. Epub 2018 Feb 20. Ecotoxicology. 2018. PMID: 29464532
-
Combining six genome scan methods to detect candidate genes to salinity in the Mediterranean striped red mullet (Mullus surmuletus).BMC Genomics. 2018 Mar 27;19(1):217. doi: 10.1186/s12864-018-4579-z. BMC Genomics. 2018. PMID: 29580201 Free PMC article.
-
Impacts of climate change on mangrove subsistence fisheries: a global review.Mar Life Sci Technol. 2024 Jun 5;6(4):610-630. doi: 10.1007/s42995-024-00231-3. eCollection 2024 Nov. Mar Life Sci Technol. 2024. PMID: 39620095 Free PMC article. Review.
-
Salinity Tolerance in Freshwater Drum (Aplodinotus grunniens): Investigating Biochemical, Antioxidant, Digestive Enzyme, and Gene Expression Responses to Acute Salinity Stress.Animals (Basel). 2025 Apr 1;15(7):1015. doi: 10.3390/ani15071015. Animals (Basel). 2025. PMID: 40218412 Free PMC article.
-
The time course of molecular acclimation to seawater in a euryhaline fish.Sci Rep. 2021 Sep 13;11(1):18127. doi: 10.1038/s41598-021-97295-3. Sci Rep. 2021. PMID: 34518569 Free PMC article.
References
-
- Bates, D. , Maechler M., Bolker B., and Walker S. 2015. Fitting linear mixed‐effects models using lme4. Journal of Statistical Software 67:1–48.
-
- Bennett, W. 2005. Critical assessment of Delta Smelt in the San Francisco Estuary, California. San Francisco Estuary and Watershed Science 3:1–72.
-
- Boeuf, G. , and Payan P. 2001. How should salinity influence fish growth? Comparative Biochemistry and Physiology Part C: Toxicology and Pharmacology 130:411–423. - PubMed
-
- Bolger, T. , and Connolly P. L. 1989. The selection of suitable indices for the measurement and analysis of fish condition. Journal of Fish Biology 34:171–182.
-
- Bradley, T. 2009. Animal Osmoregulation. Oxford University Press, New York.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

