Molecular Evolutionary Pathways toward Two Successful Community-Associated but Multidrug-Resistant ST59 Methicillin-Resistant Staphylococcus aureus Lineages in Taiwan: Dynamic Modes of Mobile Genetic Element Salvages
- PMID: 27606427
- PMCID: PMC5015870
- DOI: 10.1371/journal.pone.0162526
Molecular Evolutionary Pathways toward Two Successful Community-Associated but Multidrug-Resistant ST59 Methicillin-Resistant Staphylococcus aureus Lineages in Taiwan: Dynamic Modes of Mobile Genetic Element Salvages
Abstract
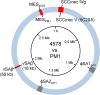
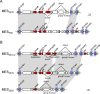
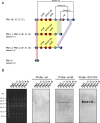
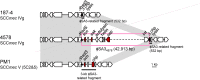
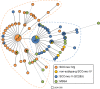
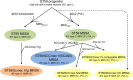
Clonal complex 59 (CC59) Staphylococcus aureus in Taiwan includes both methicillin-susceptible S. aureus (MSSA) and methicillin-resistant S. aureus (MRSA). As the most prominent community-associated MRSA (CA-MRSA) in Taiwan, CC59 has two major clones characterized as PVL-negative SCCmec IV (carrying the staphylococcal cassette chromosome mec IV but Panton-Valentine leukocidin-negative) and PVL-positive SCCmec V (5C2&5). We investigated the drug resistance, phylogeny and the distribution and sequence variation of SCCmec, staphylococcal bacteriophage φSA3, genomic island νSaβ and MES (an enterococcal mobile genetic element conferring multidrug resistance) in 195 CC59 S. aureus. Sequencing and PCR mapping revealed that all of the CC59/SCCmec V (5C2&5) MRSA strains had acquired MESPM1 or its segregants, and obtained a φSA3-related fragment in νSaβ. In contrast, MES6272-2 and MES4578, which showed gentamicin resistance that was not encoded by MESPM1, were dominant in SCCmec IVg MRSA. Translocation of a whole φSA3 into νSaβ instead of only a φSA3-related fragment was common in SCCmec IVg MRSA. However, the non-subtype-g SCCmec IV MRSA (SCCmec IVa is the major) still carried MES and νSaβ structures similar to those in SCCmec V (5C2&5) MRSA. A minimum spanning tree constructed by multiple-locus variable-number tandem repeat analysis revealed that SCCmec IVg MRSA and SCCmec V (5C2&5) MRSA grouped respectively in two major clades. The CC59 MSSA was equally distributed among the two clades, while the non-subtype-g SCCmec IV MRSA mostly clustered with SCCmec V (5C2&5) MRSA. Our findings strongly suggest that CC59 MSSA acquired divergent mobile genetic elements and evolved to SCCmec IVg MRSA and SCCmec V (5C2&5) MRSA/non-subtype-g SCCmec IV MRSA independently. The evolutionary history of CC59 S. aureus explains how mobile genetic elements increase the antimicrobial resistance and virulence and contribute to the success of CA-MRSA in Taiwan.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Tracking the evolution of the two successful CC59 methicillin-resistant Staphylococcus aureus clones in Taiwan: the divergence time of the two clades is estimated to be the 1980s.Int J Antimicrob Agents. 2020 Aug;56(2):106047. doi: 10.1016/j.ijantimicag.2020.106047. Epub 2020 Jun 13. Int J Antimicrob Agents. 2020. PMID: 32544568
-
Evolution and Population Dynamics of Clonal Complex 152 Community-Associated Methicillin-Resistant Staphylococcus aureus.mSphere. 2020 Jul 1;5(4):e00226-20. doi: 10.1128/mSphere.00226-20. mSphere. 2020. PMID: 32611695 Free PMC article.
-
Differentiation of clonal complex 59 community-associated methicillin-resistant Staphylococcus aureus in Western Australia.Antimicrob Agents Chemother. 2010 May;54(5):1914-21. doi: 10.1128/AAC.01287-09. Epub 2010 Mar 8. Antimicrob Agents Chemother. 2010. PMID: 20211891 Free PMC article.
-
The evolution of Staphylococcus aureus.Infect Genet Evol. 2008 Dec;8(6):747-63. doi: 10.1016/j.meegid.2008.07.007. Epub 2008 Jul 29. Infect Genet Evol. 2008. PMID: 18718557 Review.
-
Community-associated meticillin-resistant Staphylococcus aureus in children in Taiwan, 2000s.Int J Antimicrob Agents. 2011 Jul;38(1):2-8. doi: 10.1016/j.ijantimicag.2011.01.011. Epub 2011 Mar 11. Int J Antimicrob Agents. 2011. PMID: 21397461 Review.
Cited by
-
In Vivo Effect of Halicin on Methicillin-Resistant Staphylococcus aureus-Infected Caenorhabditis elegans and Its Clinical Potential.Antibiotics (Basel). 2024 Sep 23;13(9):906. doi: 10.3390/antibiotics13090906. Antibiotics (Basel). 2024. PMID: 39335079 Free PMC article.
-
ESKAPE in China: epidemiology and characteristics of antibiotic resistance.Emerg Microbes Infect. 2024 Dec;13(1):2317915. doi: 10.1080/22221751.2024.2317915. Epub 2024 Feb 23. Emerg Microbes Infect. 2024. PMID: 38356197 Free PMC article. Review.
-
The Genomic Context for the Evolution and Transmission of Community-Associated Staphylococcus aureus ST59 Through the Food Chain.Front Microbiol. 2020 Mar 17;11:422. doi: 10.3389/fmicb.2020.00422. eCollection 2020. Front Microbiol. 2020. PMID: 32256477 Free PMC article.
-
Staphylococcus aureus ST59: Concurrent but Separate Evolution of North American and East Asian Lineages.Front Microbiol. 2021 Feb 10;12:631845. doi: 10.3389/fmicb.2021.631845. eCollection 2021. Front Microbiol. 2021. PMID: 33643261 Free PMC article.
-
Fatal case of ST8/SCCmecIVl community-associated methicillin-resistant Staphylococcus aureus infection in Japan.New Microbes New Infect. 2018 Aug 11;26:30-36. doi: 10.1016/j.nmni.2018.08.004. eCollection 2018 Nov. New Microbes New Infect. 2018. PMID: 30245831 Free PMC article.
References
-
- Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet. 2006; 368: 874–885. - PubMed
-
- Yamamoto T, Hung WC, Takano T, Nishiyama A. Genetic nature and virulence of community-associated methicillin-resistant Staphylococcus aureus. BioMed. 2013; 3: 2–18.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

