Evolution and Ecology of Actinobacteria and Their Bioenergy Applications
- PMID: 27607553
- PMCID: PMC5703056
- DOI: 10.1146/annurev-micro-102215-095748
Evolution and Ecology of Actinobacteria and Their Bioenergy Applications
Abstract
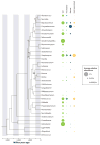
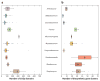
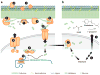
The ancient phylum Actinobacteria is composed of phylogenetically and physiologically diverse bacteria that help Earth's ecosystems function. As free-living organisms and symbionts of herbivorous animals, Actinobacteria contribute to the global carbon cycle through the breakdown of plant biomass. In addition, they mediate community dynamics as producers of small molecules with diverse biological activities. Together, the evolution of high cellulolytic ability and diverse chemistry, shaped by their ecological roles in nature, make Actinobacteria a promising group for the bioenergy industry. Specifically, their enzymes can contribute to industrial-scale breakdown of cellulosic plant biomass into simple sugars that can then be converted into biofuels. Furthermore, harnessing their ability to biosynthesize a range of small molecules has potential for the production of specialty biofuels.
Keywords: Streptomyces; actinomycetes; biofuels; biotechnology; cellulases; symbiosis.
Figures
Similar articles
-
Pharmacological Potential of Phylogenetically Diverse Actinobacteria Isolated from Deep-Sea Coral Ecosystems of the Submarine Avilés Canyon in the Cantabrian Sea.Microb Ecol. 2017 Feb;73(2):338-352. doi: 10.1007/s00248-016-0845-2. Epub 2016 Sep 10. Microb Ecol. 2017. PMID: 27614749
-
A comprehensive review on strategic study of cellulase producing marine actinobacteria for biofuel applications.Environ Res. 2022 Nov;214(Pt 3):114018. doi: 10.1016/j.envres.2022.114018. Epub 2022 Aug 9. Environ Res. 2022. PMID: 35961544 Review.
-
Exploring biodiversity for cellulosic biofuel production.Chem Biodivers. 2010 May;7(5):1086-97. doi: 10.1002/cbdv.200900314. Chem Biodivers. 2010. PMID: 20491068 Review.
-
Biodiversity, bioactive natural products and biotechnological potential of plant-associated endophytic actinobacteria.Appl Microbiol Biotechnol. 2011 Feb;89(3):457-73. doi: 10.1007/s00253-010-2923-6. Epub 2010 Oct 13. Appl Microbiol Biotechnol. 2011. PMID: 20941490 Review.
-
The realm of cellulases in biorefinery development.Crit Rev Biotechnol. 2012 Sep;32(3):187-202. doi: 10.3109/07388551.2011.595385. Epub 2011 Sep 19. Crit Rev Biotechnol. 2012. PMID: 21929293 Review.
Cited by
-
Resveratrol-mediated attenuation of superantigen-driven acute respiratory distress syndrome is mediated by microbiota in the lungs and gut.Pharmacol Res. 2021 May;167:105548. doi: 10.1016/j.phrs.2021.105548. Epub 2021 Mar 15. Pharmacol Res. 2021. PMID: 33722710 Free PMC article.
-
No Guts About It: Captivity, But Not Neophobia Phenotype, Influences the Cloacal Microbiome of House Sparrows (Passer domesticus).Integr Org Biol. 2022 Mar 11;4(1):obac010. doi: 10.1093/iob/obac010. eCollection 2022. Integr Org Biol. 2022. PMID: 35505795 Free PMC article.
-
Comparative Metagenomics Reveals Enhanced Nutrient Cycling Potential after 2 Years of Biochar Amendment in a Tropical Oxisol.Appl Environ Microbiol. 2019 May 16;85(11):e02957-18. doi: 10.1128/AEM.02957-18. Print 2019 Jun 1. Appl Environ Microbiol. 2019. PMID: 30952661 Free PMC article.
-
The antimicrobial potential of Streptomyces from insect microbiomes.Nat Commun. 2019 Jan 31;10(1):516. doi: 10.1038/s41467-019-08438-0. Nat Commun. 2019. PMID: 30705269 Free PMC article.
-
Possibly pathogenic bacteria in aerosols and foams as a result of aeration remediation in a polluted urban waterway.Folia Microbiol (Praha). 2024 Feb;69(1):235-246. doi: 10.1007/s12223-023-01096-2. Epub 2023 Sep 30. Folia Microbiol (Praha). 2024. PMID: 37777646 Free PMC article.
References
-
- Atabani AE, Silitonga AS, Badruddin IA, Mahlia TMI, Masjuki HH, Mekhilef S. A comprehensive review on biodiesel as an alternative energy resource and its characteristics. Renew Sustain Energy Rev. 2012;16:2070–93.
-
- Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451:86–89. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

