Inhibition of BET bromodomain-dependent XIAP and FLIP expression sensitizes KRAS-mutated NSCLC to pro-apoptotic agents
- PMID: 27607580
- PMCID: PMC5059870
- DOI: 10.1038/cddis.2016.271
Inhibition of BET bromodomain-dependent XIAP and FLIP expression sensitizes KRAS-mutated NSCLC to pro-apoptotic agents
Abstract
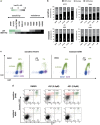
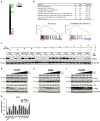
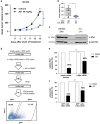
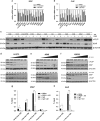
Non-small cell lung cancer (NSCLC) has the highest incidence of cancer-related death worldwide and a high medical need for more effective therapies. Small-molecule inhibitors of the bromodomain and extra terminal domain (BET) family such as JQ1, I-BET762 and OTX-015 are active in a wide range of different cancer types, including lung cancer. Although their activity on oncogene expression such as c-Myc has been addressed in many studies, the effects of BET inhibition on the apoptotic pathway remain largely unknown. Here we evaluated the activity of BET bromodomain inhibitors on cell cycle distribution and on components of the apoptosis response. Using a panel of 12 KRAS-mutated NSCLC models, we found that cell lines responsive to BET inhibitors underwent apoptosis and reduced their S-phase population, concomitant with downregulation of c-Myc expression. Conversely, ectopic c-Myc overexpression rescued the anti-proliferative effect of JQ1. In the H1373 xenograft model, treatment with JQ1 significantly reduced tumor growth and downregulated the expression of c-Myc. The effects of BET inhibition on the expression of 370 genes involved in apoptosis were compared in sensitive and resistant cells and we found the expression of the two key apoptosis regulators FLIP and XIAP to be highly BET dependent. Consistent with this, combination treatment of JQ1 with the tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) or the pro-apoptotic chemotherapeutic agent cisplatin enhanced induction of apoptosis in both BET inhibitor sensitive and resistant cells. Further we showed that combination of JQ1 with cisplatin led to significantly improved anti-tumor efficacy in A549 tumor-bearing mice. Altogether, these results show that the identification of BET-dependent genes provides guidance for the choice of drug combinations in cancer treatment. They also demonstrate that BET inhibition primes NSCLC cells for induction of apoptosis and that a combination with pro-apoptotic compounds represents a valuable strategy to overcome treatment resistance.
Conflict of interest statement
OK, RL, KAG, BH and PL are employees of Bayer Pharma AG, a company having a commercial interest in the development of BET inhibitors.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

