Condition-dependent co-regulation of genomic clusters of virulence factors in the grapevine trunk pathogen Neofusicoccum parvum
- PMID: 27608421
- PMCID: PMC6637977
- DOI: 10.1111/mpp.12491
Condition-dependent co-regulation of genomic clusters of virulence factors in the grapevine trunk pathogen Neofusicoccum parvum
Abstract
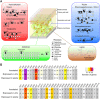
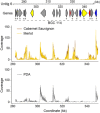
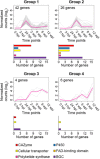
The ascomycete Neofusicoccum parvum, one of the causal agents of Botryosphaeria dieback, is a destructive wood-infecting fungus and a serious threat to grape production worldwide. The capability to colonize woody tissue, combined with the secretion of phytotoxic compounds, is thought to underlie its pathogenicity and virulence. Here, we describe the repertoire of virulence factors and their transcriptional dynamics as the fungus feeds on different substrates and colonizes the woody stem. We assembled and annotated a highly contiguous genome using single-molecule real-time DNA sequencing. Transcriptome profiling by RNA sequencing determined the genome-wide patterns of expression of virulence factors both in vitro (potato dextrose agar or medium amended with grape wood as substrate) and in planta. Pairwise statistical testing of differential expression, followed by co-expression network analysis, revealed that physically clustered genes coding for putative virulence functions were induced depending on the substrate or stage of plant infection. Co-expressed gene clusters were significantly enriched not only in genes associated with secondary metabolism, but also in those associated with cell wall degradation, suggesting that dynamic co-regulation of transcriptional networks contributes to multiple aspects of N. parvum virulence. In most of the co-expressed clusters, all genes shared at least a common motif in their promoter region, indicative of co-regulation by the same transcription factor. Co-expression analysis also identified chromatin regulators with correlated expression with inducible clusters of virulence factors, suggesting a complex, multi-layered regulation of the virulence repertoire of N. parvum.
Keywords: Botryosphaeria dieback; CAZymes; RNA-seq; cell wall degradation; secondary metabolism; single-molecule real-time (SMRT) sequencing; weighted co-expression network analysis.
© 2016 BSPP AND JOHN WILEY & SONS LTD.
Figures




Similar articles
-
Biochemical characterization of wood decay and metabolization of phenolic compounds by causal fungi of grapevine trunk diseases.PLoS One. 2025 Apr 16;20(4):e0315412. doi: 10.1371/journal.pone.0315412. eCollection 2025. PLoS One. 2025. PMID: 40238810 Free PMC article.
-
Distinctive expansion of gene families associated with plant cell wall degradation, secondary metabolism, and nutrient uptake in the genomes of grapevine trunk pathogens.BMC Genomics. 2015 Jun 19;16(1):469. doi: 10.1186/s12864-015-1624-z. BMC Genomics. 2015. PMID: 26084502 Free PMC article.
-
Global transcriptional analysis suggests Lasiodiplodia theobromae pathogenicity factors involved in modulation of grapevine defensive response.BMC Genomics. 2016 Aug 11;17(1):615. doi: 10.1186/s12864-016-2952-3. BMC Genomics. 2016. PMID: 27514986 Free PMC article.
-
Exploring Factors Conditioning the Expression of Botryosphaeria Dieback in Grapevine for Integrated Management of the Disease.Phytopathology. 2024 Jan;114(1):21-34. doi: 10.1094/PHYTO-04-23-0136-RVW. Epub 2024 Feb 14. Phytopathology. 2024. PMID: 37505093 Review.
-
Transcription factor control of virulence in phytopathogenic fungi.Mol Plant Pathol. 2021 Jul;22(7):858-881. doi: 10.1111/mpp.13056. Epub 2021 May 11. Mol Plant Pathol. 2021. PMID: 33973705 Free PMC article. Review.
Cited by
-
Diversity and bioactivities of fungal endophytes from Distylium chinense, a rare waterlogging tolerant plant endemic to the Three Gorges Reservoir.BMC Microbiol. 2019 Dec 10;19(1):278. doi: 10.1186/s12866-019-1634-0. BMC Microbiol. 2019. PMID: 31822262 Free PMC article.
-
Insights of the Neofusicoccum parvum-Liquidambar styraciflua Interaction and Identification of New Cysteine-Rich Proteins in Both Species.J Fungi (Basel). 2021 Nov 30;7(12):1027. doi: 10.3390/jof7121027. J Fungi (Basel). 2021. PMID: 34947009 Free PMC article.
-
Biochemical characterization of wood decay and metabolization of phenolic compounds by causal fungi of grapevine trunk diseases.PLoS One. 2025 Apr 16;20(4):e0315412. doi: 10.1371/journal.pone.0315412. eCollection 2025. PLoS One. 2025. PMID: 40238810 Free PMC article.
-
Comparative Transcriptomics and Gene Knockout Reveal Virulence Factors of Neofusicoccum parvum in Walnut.Front Microbiol. 2022 Jul 15;13:926620. doi: 10.3389/fmicb.2022.926620. eCollection 2022. Front Microbiol. 2022. PMID: 35910616 Free PMC article.
-
Whole-Genome Resequencing and Pan-Transcriptome Reconstruction Highlight the Impact of Genomic Structural Variation on Secondary Metabolite Gene Clusters in the Grapevine Esca Pathogen Phaeoacremonium minimum.Front Microbiol. 2018 Aug 13;9:1784. doi: 10.3389/fmicb.2018.01784. eCollection 2018. Front Microbiol. 2018. PMID: 30150972 Free PMC article.
References
-
- Abou‐Mansour, E. , Débieux, J.L. , Ramírez‐Suero, M. , Bénard‐Gellon, M. , Magnin‐Robert, M. , Spagnolo, A. , Chong, J. , Farine, S. , Bertsch, C. , L'haridon, F. , Serrano, M. , Fontaine, F. , Rego, C. and Larignon, P. (2015) Phytotoxic metabolites from Neofusicoccum parvum, a pathogen of Botryosphaeria dieback of grapevine. Phytochemistry, 115, 207–215. - PubMed
-
- Aro, N. , Pakula, T. and Penttilä, M. (2005) Transcriptional regulation of plant cell wall degradation by filamentous fungi. FEMS Microbiol. Rev. 29, 719–739. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

