Evidence for the presence of African swine fever virus in an endemic region of Western Kenya in the absence of any reported outbreak
- PMID: 27608711
- PMCID: PMC5016997
- DOI: 10.1186/s12917-016-0830-5
Evidence for the presence of African swine fever virus in an endemic region of Western Kenya in the absence of any reported outbreak
Abstract
Background: African swine fever (ASF), caused by African swine fever virus (ASFV), is a severe haemorrhagic disease of pigs, outbreaks of which can have a devastating impact upon commercial and small-holder pig production. Pig production in western Kenya is characterised by low-input, free-range systems practised by poor farmers keeping between two and ten pigs. These farmers are particularly vulnerable to the catastrophic loss of livestock assets experienced in an ASF outbreak. This study wished to expand our understanding of ASFV epidemiology during a period when no outbreaks were reported.
Results: Two hundred and seventy six whole blood samples were analysed using two independent conventional and real time PCR assays to detect ASFV. Despite no recorded outbreak of clinical ASF during this time, virus was detected in 90/277 samples analysed by conventional PCR and 142/209 samples analysed by qPCR. Genotyping of a sub-set of these samples indicated that the viruses associated with the positive samples were classified within genotype IX and that these strains were therefore genetically similar to the virus associated with the 2006/2007 ASF outbreaks in Kenya.
Conclusion: The detection of ASFV viral DNA in a relatively high number of pigs delivered for slaughter during a period with no reported outbreaks provides support for two hypotheses, which are not mutually exclusive: (1) that virus prevalence may be over-estimated by slaughter-slab sampling, relative to that prevailing in the wider pig population; (2) that sub-clinical, chronically infected or recovered pigs may be responsible for persistence of the virus in endemic areas.
Keywords: ASFV real time PCR; African swine fever virus; Epidemiology; Genotype IX; Kenya; Slaughter house; p72 PCR.
Figures
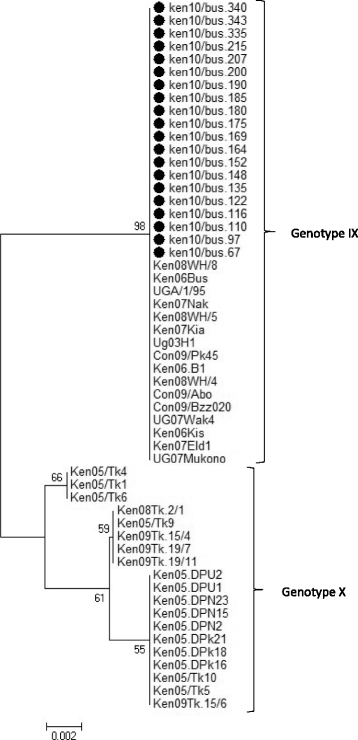
 Indicates the 20 nucleotide sequences analyzed in this study in comparison to 35 reference sequences obtained from Genbank. The 20 sequences clustered within ASFV genotype IX. The evolutionary history was inferred using the Minimum Evolution (ME) method after initial application of the Neighbor-joining algorithm. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The p-distance method was used to compute evolutionary distances and the Close-Neighbor-Interchange (CNI) algorithm at a search level of 1 was used to determine the strength of the ME tree
Indicates the 20 nucleotide sequences analyzed in this study in comparison to 35 reference sequences obtained from Genbank. The 20 sequences clustered within ASFV genotype IX. The evolutionary history was inferred using the Minimum Evolution (ME) method after initial application of the Neighbor-joining algorithm. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The p-distance method was used to compute evolutionary distances and the Close-Neighbor-Interchange (CNI) algorithm at a search level of 1 was used to determine the strength of the ME tree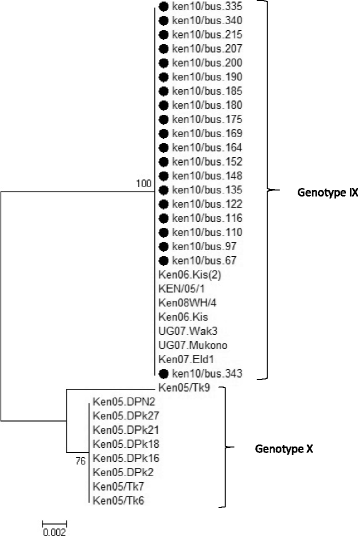
 Indicates the 20 sequences analyzed in this study that cluster within genotype IX in comparison to 16 reference sequences obtained from Genbank. The evolutionary history was inferred using the Minimum Evolution method after initial utilization of the Neighbor-joining algorithm. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method and are in the units of the number of base differences per site. The ME tree was searched using the Close-Neighbour-Interchange (CNI) algorithm at a search level of 1
Indicates the 20 sequences analyzed in this study that cluster within genotype IX in comparison to 16 reference sequences obtained from Genbank. The evolutionary history was inferred using the Minimum Evolution method after initial utilization of the Neighbor-joining algorithm. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method and are in the units of the number of base differences per site. The ME tree was searched using the Close-Neighbour-Interchange (CNI) algorithm at a search level of 1Similar articles
-
Detection of African swine fever virus in the tissues of asymptomatic pigs in smallholder farming systems along the Kenya-Uganda border: implications for transmission in endemic areas and ASF surveillance in East Africa.J Gen Virol. 2017 Jul;98(7):1806-1814. doi: 10.1099/jgv.0.000848. Epub 2017 Jul 10. J Gen Virol. 2017. PMID: 28721858
-
Epidemiology and ecology of the sylvatic cycle of African Swine Fever Virus in Kenya.Virus Res. 2024 Oct;348:199434. doi: 10.1016/j.virusres.2024.199434. Epub 2024 Jul 19. Virus Res. 2024. PMID: 39004284 Free PMC article.
-
Genetic profile of African swine fever virus responsible for the 2019 outbreak in northern Malawi.BMC Vet Res. 2020 Aug 28;16(1):316. doi: 10.1186/s12917-020-02536-8. BMC Vet Res. 2020. PMID: 32859205 Free PMC article.
-
African Swine Fever Virus Circulation between Tanzania and Neighboring Countries: A Systematic Review and Meta-Analysis.Viruses. 2021 Feb 15;13(2):306. doi: 10.3390/v13020306. Viruses. 2021. PMID: 33672090 Free PMC article.
-
[African swine fever in Russian Federation].Vopr Virusol. 2012 Sep-Oct;57(5):4-10. Vopr Virusol. 2012. PMID: 23248852 Review. Russian.
Cited by
-
Piloting the effectiveness of pig health education in combination with oxfendazole treatment on prevention and/or control of porcine cysticercosis, gastrointestinal parasites, African swine fever and ectoparasites in Angónia District, Mozambique.Trop Anim Health Prod. 2018 Mar;50(3):589-601. doi: 10.1007/s11250-017-1474-6. Epub 2017 Nov 14. Trop Anim Health Prod. 2018. PMID: 29139069
-
Thoughts on African Swine Fever Vaccines.Viruses. 2021 May 20;13(5):943. doi: 10.3390/v13050943. Viruses. 2021. PMID: 34065425 Free PMC article.
-
African Local Pig Genetic Resources in the Context of Climate Change Adaptation.Animals (Basel). 2024 Aug 19;14(16):2407. doi: 10.3390/ani14162407. Animals (Basel). 2024. PMID: 39199941 Free PMC article. Review.
-
African swine fever: Etiology, epidemiological status in Korea, and perspective on control.J Vet Sci. 2020 Mar;21(2):e38. doi: 10.4142/jvs.2020.21.e38. J Vet Sci. 2020. PMID: 32233141 Free PMC article. Review.
-
Phylodynamics and evolutionary epidemiology of African swine fever p72-CVR genes in Eurasia and Africa.PLoS One. 2018 Feb 28;13(2):e0192565. doi: 10.1371/journal.pone.0192565. eCollection 2018. PLoS One. 2018. PMID: 29489860 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

