HMGB4 is expressed by neuronal cells and affects the expression of genes involved in neural differentiation
- PMID: 27608812
- PMCID: PMC5036535
- DOI: 10.1038/srep32960
HMGB4 is expressed by neuronal cells and affects the expression of genes involved in neural differentiation
Abstract
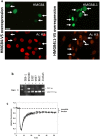
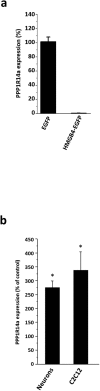
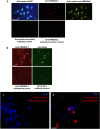
HMGB4 is a new member in the family of HMGB proteins that has been characterized in sperm cells, but little is known about its functions in somatic cells. Here we show that HMGB4 and the highly similar rat Transition Protein 4 (HMGB4L1) are expressed in neuronal cells. Both proteins had slow mobility in nucleus of living NIH-3T3 cells. They interacted with histones and their differential expression in transformed cells of the nervous system altered the post-translational modification statuses of histones in vitro. Overexpression of HMGB4 in HEK 293T cells made cells more susceptible to cell death induced by topoisomerase inhibitors in an oncology drug screening array and altered variant composition of histone H3. HMGB4 regulated over 800 genes in HEK 293T cells with a p-value ≤0.013 (n = 3) in a microarray analysis and displayed strongest association with adhesion and histone H2A -processes. In neuronal and transformed cells HMGB4 regulated the expression of an oligodendrocyte marker gene PPP1R14a and other neuronal differentiation marker genes. In conclusion, our data suggests that HMGB4 is a factor that regulates chromatin and expression of neuronal differentiation markers.
Figures



Similar articles
-
HMGB4, a novel member of the HMGB family, is preferentially expressed in the mouse testis and localizes to the basal pole of elongating spermatids.Biol Reprod. 2009 Feb;80(2):358-66. doi: 10.1095/biolreprod.108.070243. Epub 2008 Nov 5. Biol Reprod. 2009. PMID: 18987332
-
Nucleocytoplasmic distribution of the Arabidopsis chromatin-associated HMGB2/3 and HMGB4 proteins.Plant Physiol. 2010 Dec;154(4):1831-41. doi: 10.1104/pp.110.163055. Epub 2010 Oct 12. Plant Physiol. 2010. PMID: 20940346 Free PMC article.
-
HMGB proteins: interactions with DNA and chromatin.Biochim Biophys Acta. 2010 Jan-Feb;1799(1-2):101-13. doi: 10.1016/j.bbagrm.2009.09.008. Biochim Biophys Acta. 2010. PMID: 20123072 Review.
-
Interaction of maize chromatin-associated HMG proteins with mononucleosomes: role of core and linker histones.Biol Chem. 2003 Jul;384(7):1019-27. doi: 10.1515/BC.2003.114. Biol Chem. 2003. PMID: 12956418
-
HMGB proteins and transcriptional regulation.Biochim Biophys Acta. 2010 Jan-Feb;1799(1-2):114-8. doi: 10.1016/j.bbagrm.2009.11.005. Biochim Biophys Acta. 2010. PMID: 20123073 Review.
Cited by
-
Oligodendroglial Epigenetics, from Lineage Specification to Activity-Dependent Myelination.Life (Basel). 2021 Jan 15;11(1):62. doi: 10.3390/life11010062. Life (Basel). 2021. PMID: 33467699 Free PMC article. Review.
-
HMGB1/RAGE axis in tumor development: unraveling its significance.Front Oncol. 2024 Mar 1;14:1336191. doi: 10.3389/fonc.2024.1336191. eCollection 2024. Front Oncol. 2024. PMID: 38529373 Free PMC article. Review.
-
Pathophysiological role of high mobility group box-1 signaling in neurodegenerative diseases.Inflammopharmacology. 2025 Feb;33(2):703-727. doi: 10.1007/s10787-024-01595-9. Epub 2024 Nov 15. Inflammopharmacology. 2025. PMID: 39546221 Review.
-
Structure and Functions of HMGB3 Protein.Int J Mol Sci. 2024 Jul 12;25(14):7656. doi: 10.3390/ijms25147656. Int J Mol Sci. 2024. PMID: 39062899 Free PMC article. Review.
-
Whole-genome resequencing of Japanese whiting ( Sillago japonica) provide insights into local adaptations.Zool Res. 2021 Sep 18;42(5):548-561. doi: 10.24272/j.issn.2095-8137.2021.116. Zool Res. 2021. PMID: 34327887 Free PMC article.
References
-
- Gaillard C. & Strauss F. Association of poly(CA).poly(TG) DNA fragments into four-stranded complexes bound by HMG1 and 2. Science 264, 433–436 (1994). - PubMed
-
- Catena R. et al. HMGB4, a novel member of the HMGB family, is preferentially expressed in the mouse testis and localizes to the basal pole of elongating spermatids. Biol. Reprod. 80, 358–366 (2009). - PubMed
-
- Zhou Q. et al. A multipotent progenitor domain guides pancreatic organogenesis. Dev. Cell. 13, 103–114 (2007). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

