Single-molecule Imaging Analysis of Binding, Processive Movement, and Dissociation of Cellobiohydrolase Trichoderma reesei Cel6A and Its Domains on Crystalline Cellulose
- PMID: 27609516
- PMCID: PMC5077181
- DOI: 10.1074/jbc.M116.752048
Single-molecule Imaging Analysis of Binding, Processive Movement, and Dissociation of Cellobiohydrolase Trichoderma reesei Cel6A and Its Domains on Crystalline Cellulose
Abstract
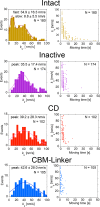
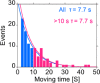
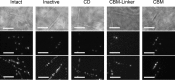
Trichoderma reesei Cel6A (TrCel6A) is a cellobiohydrolase that hydrolyzes crystalline cellulose into cellobiose. Here we directly observed the reaction cycle (binding, surface movement, and dissociation) of single-molecule intact TrCel6A, isolated catalytic domain (CD), cellulose-binding module (CBM), and CBM and linker (CBM-linker) on crystalline cellulose Iα The CBM-linker showed a binding rate constant almost half that of intact TrCel6A, whereas those of the CD and CBM were only one-tenth of intact TrCel6A. These results indicate that the glycosylated linker region largely contributes to initial binding on crystalline cellulose. After binding, all samples showed slow and fast dissociations, likely caused by the two different bound states due to the heterogeneity of cellulose surface. The CBM showed much higher specificity to the high affinity site than to the low affinity site, whereas the CD did not, suggesting that the CBM leads the CD to the hydrophobic surface of crystalline cellulose. On the cellulose surface, intact molecules showed slow processive movements (8.8 ± 5.5 nm/s) and fast diffusional movements (30-40 nm/s), whereas the CBM-Linker, CD, and a catalytically inactive full-length mutant showed only fast diffusional movements. These results suggest that both direct binding and surface diffusion contribute to searching of the hydrolysable point of cellulose chains. The duration time constant for the processive movement was 7.7 s, and processivity was estimated as 68 ± 42. Our results reveal the role of each domain in the elementary steps of the reaction cycle and provide the first direct evidence of the processive movement of TrCel6A on crystalline cellulose.
Keywords: cellulase; cellulose; enzyme kinetics; enzyme mechanism; microscopic imaging; molecular motor; processivity; protein domain; single-molecule biophysics.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





Similar articles
-
Domain architecture divergence leads to functional divergence in binding and catalytic domains of bacterial and fungal cellobiohydrolases.J Biol Chem. 2020 Oct 23;295(43):14606-14617. doi: 10.1074/jbc.RA120.014792. Epub 2020 Aug 18. J Biol Chem. 2020. PMID: 32816991 Free PMC article.
-
Rate-limiting step and substrate accessibility of cellobiohydrolase Cel6A from Trichoderma reesei.FEBS J. 2018 Dec;285(23):4482-4493. doi: 10.1111/febs.14668. Epub 2018 Oct 17. FEBS J. 2018. PMID: 30281909
-
The tryptophan residue at the active site tunnel entrance of Trichoderma reesei cellobiohydrolase Cel7A is important for initiation of degradation of crystalline cellulose.J Biol Chem. 2013 May 10;288(19):13503-10. doi: 10.1074/jbc.M113.452623. Epub 2013 Mar 26. J Biol Chem. 2013. PMID: 23532843 Free PMC article.
-
High speed atomic force microscopy visualizes processive movement of Trichoderma reesei cellobiohydrolase I on crystalline cellulose.J Biol Chem. 2009 Dec 25;284(52):36186-36190. doi: 10.1074/jbc.M109.034611. Epub 2009 Oct 26. J Biol Chem. 2009. PMID: 19858200 Free PMC article.
-
Inter-domain Synergism Is Required for Efficient Feeding of Cellulose Chain into Active Site of Cellobiohydrolase Cel7A.J Biol Chem. 2016 Dec 9;291(50):26013-26023. doi: 10.1074/jbc.M116.756007. Epub 2016 Oct 25. J Biol Chem. 2016. PMID: 27780868 Free PMC article.
Cited by
-
Are cellulases slow? Kinetic and thermodynamic limitations for enzymatic breakdown of cellulose.BBA Adv. 2024 Dec 6;7:100128. doi: 10.1016/j.bbadva.2024.100128. eCollection 2025. BBA Adv. 2024. PMID: 39758504 Free PMC article. Review.
-
Novel Endotype Xanthanase from Xanthan-Degrading Microbacterium sp. Strain XT11.Appl Environ Microbiol. 2019 Jan 9;85(2):e01800-18. doi: 10.1128/AEM.01800-18. Print 2019 Jan 15. Appl Environ Microbiol. 2019. PMID: 30413476 Free PMC article.
-
Thermotolerance Mechanism of Fungal GH6 Cellobiohydrolase. Part II. Structural Analysis of Thermotolerant Mutant from the Basidiomycete Phanerochaete chrysosporium.J Appl Glycosci (1999). 2024 May 20;71(2):63-72. doi: 10.5458/jag.jag.JAG-2023_0018. eCollection 2024. J Appl Glycosci (1999). 2024. PMID: 38863950 Free PMC article.
-
Acoustic force spectroscopy reveals subtle differences in cellulose unbinding behavior of carbohydrate-binding modules.Proc Natl Acad Sci U S A. 2022 Oct 18;119(42):e2117467119. doi: 10.1073/pnas.2117467119. Epub 2022 Oct 10. Proc Natl Acad Sci U S A. 2022. PMID: 36215467 Free PMC article.
-
Thermobifida fusca Cel6B moves bidirectionally while processively degrading cellulose.Biotechnol Biofuels Bioprod. 2024 Dec 4;17(1):140. doi: 10.1186/s13068-024-02588-0. Biotechnol Biofuels Bioprod. 2024. PMID: 39633461 Free PMC article.
References
-
- Himmel M. E., Ding S. Y., Johnson D. K., Adney W. S., Nimlos M. R., Brady J. W., and Foust T. D. (2007) Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315, 804–807 - PubMed
-
- Chundawat S. P., Beckham G. T., Himmel M. E., and Dale B. E. (2011) Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu. Rev. Chem. Biomol. Eng. 2, 121–145 - PubMed
-
- Payne C. M., Knott B. C., Mayes H. B., Hansson H., Himmel M. E., Sandgren M., Ståhlberg J., and Beckham G. T. (2015) Fungal cellulases. Chem. Rev. 115, 1308–1448 - PubMed
-
- Wilson D. B. (2011) Microbial diversity of cellulose hydrolysis. Curr. Opin. Microbiol. 14, 259–263 - PubMed
-
- Imai T., Boisset C., Samejima M., Igarashi K., and Sugiyama J. (1998) Unidirectional processive action of cellobiohydrolase Cel7A on Valonia cellulose microcrystals. FEBS Lett. 432, 113–116 - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

