Single-molecule Imaging Analysis of Binding, Processive Movement, and Dissociation of Cellobiohydrolase Trichoderma reesei Cel6A and Its Domains on Crystalline Cellulose
- PMID: 27609516
- PMCID: PMC5077181
- DOI: 10.1074/jbc.M116.752048
Single-molecule Imaging Analysis of Binding, Processive Movement, and Dissociation of Cellobiohydrolase Trichoderma reesei Cel6A and Its Domains on Crystalline Cellulose
Abstract
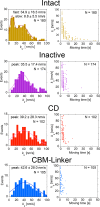
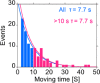
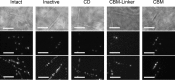
Trichoderma reesei Cel6A (TrCel6A) is a cellobiohydrolase that hydrolyzes crystalline cellulose into cellobiose. Here we directly observed the reaction cycle (binding, surface movement, and dissociation) of single-molecule intact TrCel6A, isolated catalytic domain (CD), cellulose-binding module (CBM), and CBM and linker (CBM-linker) on crystalline cellulose Iα The CBM-linker showed a binding rate constant almost half that of intact TrCel6A, whereas those of the CD and CBM were only one-tenth of intact TrCel6A. These results indicate that the glycosylated linker region largely contributes to initial binding on crystalline cellulose. After binding, all samples showed slow and fast dissociations, likely caused by the two different bound states due to the heterogeneity of cellulose surface. The CBM showed much higher specificity to the high affinity site than to the low affinity site, whereas the CD did not, suggesting that the CBM leads the CD to the hydrophobic surface of crystalline cellulose. On the cellulose surface, intact molecules showed slow processive movements (8.8 ± 5.5 nm/s) and fast diffusional movements (30-40 nm/s), whereas the CBM-Linker, CD, and a catalytically inactive full-length mutant showed only fast diffusional movements. These results suggest that both direct binding and surface diffusion contribute to searching of the hydrolysable point of cellulose chains. The duration time constant for the processive movement was 7.7 s, and processivity was estimated as 68 ± 42. Our results reveal the role of each domain in the elementary steps of the reaction cycle and provide the first direct evidence of the processive movement of TrCel6A on crystalline cellulose.
Keywords: cellulase; cellulose; enzyme kinetics; enzyme mechanism; microscopic imaging; molecular motor; processivity; protein domain; single-molecule biophysics.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
-
- Himmel M. E., Ding S. Y., Johnson D. K., Adney W. S., Nimlos M. R., Brady J. W., and Foust T. D. (2007) Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315, 804–807 - PubMed
-
- Chundawat S. P., Beckham G. T., Himmel M. E., and Dale B. E. (2011) Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu. Rev. Chem. Biomol. Eng. 2, 121–145 - PubMed
-
- Payne C. M., Knott B. C., Mayes H. B., Hansson H., Himmel M. E., Sandgren M., Ståhlberg J., and Beckham G. T. (2015) Fungal cellulases. Chem. Rev. 115, 1308–1448 - PubMed
-
- Wilson D. B. (2011) Microbial diversity of cellulose hydrolysis. Curr. Opin. Microbiol. 14, 259–263 - PubMed
-
- Imai T., Boisset C., Samejima M., Igarashi K., and Sugiyama J. (1998) Unidirectional processive action of cellobiohydrolase Cel7A on Valonia cellulose microcrystals. FEBS Lett. 432, 113–116 - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

