tRNA-derived short RNAs bind to Saccharomyces cerevisiae ribosomes in a stress-dependent manner and inhibit protein synthesis in vitro
- PMID: 27609601
- PMCID: PMC5049586
- DOI: 10.1093/femsyr/fow077
tRNA-derived short RNAs bind to Saccharomyces cerevisiae ribosomes in a stress-dependent manner and inhibit protein synthesis in vitro
Abstract
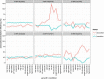
Recently, a number of ribosome-associated non-coding RNAs (rancRNAs) have been discovered in all three domains of life. In our previous studies, we have described several types of rancRNAs in Saccharomyces cerevisiae, derived from many cellular RNAs, including mRNAs, rRNAs, tRNAs and snoRNAs. Here, we present the evidence that the tRNA fragments from simple eukaryotic organism S. cerevisiae directly bind to the ribosomes. Interestingly, rancRNA-tRFs in yeast are derived from both, 5'- and 3'-part of the tRNAs and both types of tRFs associate with the ribosomes in vitro The location of tRFs within the ribosomes is distinct from classical A- and P-tRNA binding sites. Moreover, 3'-tRFs bind to the distinct site than 5'-tRFs. These interactions are stress dependent and as a consequence, provoke regulation of protein biosynthesis. We observe strong correlation between tRF binding to the ribosomes and inhibition of protein biosynthesis in particular environmental conditions. These results implicate the existence of an ancient and conserved mechanism of translation regulation with the involvement of ribosome-associating tRNA-derived fragments.
Keywords: Saccharomyces cerevisiae; protein biosynthesis regulation; rancRNAs; ribosome; tRFs; tRNA-derived small RNAs.
© FEMS 2016.
Figures
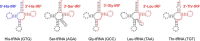




References
-
- Bąkowska-Żywicka K, Kietrys AM, Twardowski T. Antisense oligonucleotides targeting universally conserved 26S rRNA domains of plant ribosomes at different steps of polypeptide elongation. Oligonucleotides. 2008;18:175–86. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

