Development of a generally applicable morphokinetic algorithm capable of predicting the implantation potential of embryos transferred on Day 3
- PMID: 27609980
- PMCID: PMC5027927
- DOI: 10.1093/humrep/dew188
Development of a generally applicable morphokinetic algorithm capable of predicting the implantation potential of embryos transferred on Day 3
Abstract
Study question: Can a generally applicable morphokinetic algorithm suitable for Day 3 transfers of time-lapse monitored embryos originating from different culture conditions and fertilization methods be developed for the purpose of supporting the embryologist's decision on which embryo to transfer back to the patient in assisted reproduction?
Summary answer: The algorithm presented here can be used independently of culture conditions and fertilization method and provides predictive power not surpassed by other published algorithms for ranking embryos according to their blastocyst formation potential.
What is known already: Generally applicable algorithms have so far been developed only for predicting blastocyst formation. A number of clinics have reported validated implantation prediction algorithms, which have been developed based on clinic-specific culture conditions and clinical environment. However, a generally applicable embryo evaluation algorithm based on actual implantation outcome has not yet been reported.
Study design, size, duration: Retrospective evaluation of data extracted from a database of known implantation data (KID) originating from 3275 embryos transferred on Day 3 conducted in 24 clinics between 2009 and 2014. The data represented different culture conditions (reduced and ambient oxygen with various culture medium strategies) and fertilization methods (IVF, ICSI). The capability to predict blastocyst formation was evaluated on an independent set of morphokinetic data from 11 218 embryos which had been cultured to Day 5. PARTICIPANTS/MATERIALS, SETTING,
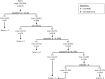
Methods: The algorithm was developed by applying automated recursive partitioning to a large number of annotation types and derived equations, progressing to a five-fold cross-validation test of the complete data set and a validation test of different incubation conditions and fertilization methods. The results were expressed as receiver operating characteristics curves using the area under the curve (AUC) to establish the predictive strength of the algorithm.
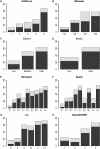
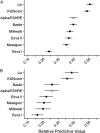
Main results and the role of chance: By applying the here developed algorithm (KIDScore), which was based on six annotations (the number of pronuclei equals 2 at the 1-cell stage, time from insemination to pronuclei fading at the 1-cell stage, time from insemination to the 2-cell stage, time from insemination to the 3-cell stage, time from insemination to the 5-cell stage and time from insemination to the 8-cell stage) and ranking the embryos in five groups, the implantation potential of the embryos was predicted with an AUC of 0.650. On Day 3 the KIDScore algorithm was capable of predicting blastocyst development with an AUC of 0.745 and blastocyst quality with an AUC of 0.679. In a comparison of blastocyst prediction including six other published algorithms and KIDScore, only KIDScore and one more algorithm surpassed an algorithm constructed on conventional Alpha/ESHRE consensus timings in terms of predictive power.
Limitations, reasons for caution: Some morphological assessments were not available and consequently three of the algorithms in the comparison were not used in full and may therefore have been put at a disadvantage. Algorithms based on implantation data from Day 3 embryo transfers require adjustments to be capable of predicting the implantation potential of Day 5 embryo transfers. The current study is restricted by its retrospective nature and absence of live birth information. Prospective Randomized Controlled Trials should be used in future studies to establish the value of time-lapse technology and morphokinetic evaluation.
Wider implications of the findings: Algorithms applicable to different culture conditions can be developed if based on large data sets of heterogeneous origin.
Study funding/competing interests: This study was funded by Vitrolife A/S, Denmark and Vitrolife AB, Sweden. B.M.P.'s company BMP Analytics is performing consultancy for Vitrolife A/S. M.B. is employed at Vitrolife A/S. M.M.'s company ilabcomm GmbH received honorarium for consultancy from Vitrolife AB. D.K.G. received research support from Vitrolife AB.
Keywords: algorithm; blastocyst; decision support tool; embryo selection; implantation; morphokinetic; prediction model; time-lapse.
© The Author 2016. Published by Oxford University Press on behalf of the European Society of Human Reproduction and Embryology.
Figures
Similar articles
-
Clinical validation of an automatic classification algorithm applied on cleavage stage embryos: analysis for blastulation, euploidy, implantation, and live-birth potential.Hum Reprod. 2023 Jun 1;38(6):1060-1075. doi: 10.1093/humrep/dead058. Hum Reprod. 2023. PMID: 37018626
-
Correlation between aneuploidy, standard morphology evaluation and morphokinetic development in 1730 biopsied blastocysts: a consecutive case series study.Hum Reprod. 2016 Oct;31(10):2245-54. doi: 10.1093/humrep/dew183. Epub 2016 Sep 2. Hum Reprod. 2016. PMID: 27591227
-
Time-lapse deselection model for human day 3 in vitro fertilization embryos: the combination of qualitative and quantitative measures of embryo growth.Fertil Steril. 2016 Mar;105(3):656-662.e1. doi: 10.1016/j.fertnstert.2015.11.003. Epub 2015 Nov 23. Fertil Steril. 2016. PMID: 26616439
-
Morphological and morphokinetic associations with aneuploidy: a systematic review and meta-analysis.Hum Reprod Update. 2022 Aug 25;28(5):656-686. doi: 10.1093/humupd/dmac022. Hum Reprod Update. 2022. PMID: 35613016
-
Is it time to reconsider how to manage oocytes affected by smooth endoplasmic reticulum aggregates?Hum Reprod. 2019 Apr 1;34(4):591-600. doi: 10.1093/humrep/dez010. Hum Reprod. 2019. PMID: 30805638 Review.
Cited by
-
A novel embryo quality scoring system to compare groups of embryos at different developmental stages.J Assist Reprod Genet. 2021 May;38(5):1123-1132. doi: 10.1007/s10815-021-02117-0. Epub 2021 Mar 1. J Assist Reprod Genet. 2021. PMID: 33646470 Free PMC article.
-
Enhancing predictive models for egg donation: time to blastocyst hatching and machine learning insights.Reprod Biol Endocrinol. 2024 Sep 11;22(1):116. doi: 10.1186/s12958-024-01285-9. Reprod Biol Endocrinol. 2024. PMID: 39261843 Free PMC article.
-
Pseudo contrastive labeling for predicting IVF embryo developmental potential.Sci Rep. 2022 Feb 15;12(1):2488. doi: 10.1038/s41598-022-06336-y. Sci Rep. 2022. PMID: 35169194 Free PMC article.
-
Asynchronous division at 4-8-cell stage of preimplantation embryos affects live birth through ICM/TE differentiation.Sci Rep. 2022 Jun 7;12(1):9411. doi: 10.1038/s41598-022-13646-8. Sci Rep. 2022. PMID: 35672442 Free PMC article.
-
Time-lapse technology for embryo culture and selection.Ups J Med Sci. 2020 May;125(2):77-84. doi: 10.1080/03009734.2020.1728444. Epub 2020 Feb 25. Ups J Med Sci. 2020. PMID: 32096675 Free PMC article. Review.
References
-
- Adamson GD, Abusief ME, Palao L, Witmer J, Palao LM, Gvakharia M. Improved implantation rates of day 3 embryo transfers with the use of an automated time-lapse-enabled test to aid in embryo selection. Fertil Steril 2016;105:369–375. - PubMed
-
- Adashi EY, Barri PN, Berkowitz R, Braude P, Bryan E, Carr J, Cohen J, Collins J, Devroey P, Frydman R et al. . Infertility therapy-associated multiple pregnancies (births): an ongoing epidemic. Reprod Biomed Online 2003;7:515–542. - PubMed
-
- Aguilar J, Motato Y, Escribá MJ, Ojeda M, Muñoz E, Meseguer M. The human first cell cycle: impact on implantation. Reprod Biomed Online 2014;28:475–484. - PubMed
-
- Aguilar J, Rubio I, Muñoz E, Pellicer A, Meseguer M. Study of nucleation status in the second cell cycle of human embryo and its impact on implantation rate. Fertil Steril 2016. doi:10.1016/j.fertnstert.2016.03.036. (Epub ahead of print). - DOI - PubMed
-
- Ahlström A, Westin C, Reismer E, Wikland M, Hardarson T. Trophectoderm morphology: an important parameter for predicting live birth after single blastocyst transfer. Hum Reprod 2011;26:3289–3296. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

