Hepatic Differentiation of Human Induced Pluripotent Stem Cells in a Perfused Three-Dimensional Multicompartment Bioreactor
- PMID: 27610270
- PMCID: PMC5003005
- DOI: 10.1089/biores.2016.0027
Hepatic Differentiation of Human Induced Pluripotent Stem Cells in a Perfused Three-Dimensional Multicompartment Bioreactor
Abstract
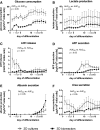
The hepatic differentiation of human induced pluripotent stem cells (hiPSC) holds great potential for application in regenerative medicine, pharmacological drug screening, and toxicity testing. However, full maturation of hiPSC into functional hepatocytes has not yet been achieved. In this study, we investigated the potential of a dynamic three-dimensional (3D) hollow fiber membrane bioreactor technology to improve the hepatic differentiation of hiPSC in comparison to static two-dimensional (2D) cultures. A total of 100 × 10(6) hiPSC were seeded into each 3D bioreactor (n = 3). Differentiation into definitive endoderm (DE) was induced by adding activin A, Wnt3a, and sodium butyrate to the culture medium. For further maturation, hepatocyte growth factor and oncostatin M were added. The same differentiation protocol was applied to hiPSC maintained in 2D cultures. Secretion of alpha-fetoprotein (AFP), a marker for DE, was significantly (p < 0.05) higher in 2D cultures, while secretion of albumin, a typical characteristic for mature hepatocytes, was higher after hepatic differentiation of hiPSC in 3D bioreactors. Functional analysis of multiple cytochrome P450 (CYP) isoenzymes showed activity of CYP1A2, CYP2B6, and CYP3A4 in both groups, although at a lower level compared to primary human hepatocytes (PHH). CYP2B6 activities were significantly (p < 0.05) higher in 3D bioreactors compared with 2D cultures, which is in line with results from gene expression. Immunofluorescence staining showed that the majority of cells was positive for albumin, cytokeratin 18 (CK18), and hepatocyte nuclear factor 4-alpha (HNF4A) at the end of the differentiation process. In addition, cytokeratin 19 (CK19) staining revealed the formation of bile duct-like structures in 3D bioreactors similar to native liver tissue. The results indicate a better maturation of hiPSC in the 3D bioreactor system compared to 2D cultures and emphasize the potential of dynamic 3D culture systems in stem cell differentiation approaches for improved formation of differentiated tissue structures.
Keywords: stem cells; tissue engineering.
Figures



References
-
- Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3:711–715 - PubMed
-
- Giri S, Bader A. Improved preclinical safety assessment using micro-BAL devices: the potential impact on human discovery and drug attrition. Drug Discov Today. 2011;16:382–397 - PubMed
-
- Ingelman-Sundberg M. Pharmacogenetics of cytochrome P450 and its applications in drug therapy: the past, present and future. Trends Pharmacol Sci. 2004;25:193–200 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
