Sequential Immunization Elicits Broadly Neutralizing Anti-HIV-1 Antibodies in Ig Knockin Mice
- PMID: 27610569
- PMCID: PMC5019122
- DOI: 10.1016/j.cell.2016.07.030
Sequential Immunization Elicits Broadly Neutralizing Anti-HIV-1 Antibodies in Ig Knockin Mice
Abstract
A vaccine that elicits broadly neutralizing antibodies (bNAbs) against HIV-1 is likely to be protective, but this has not been achieved. To explore immunization regimens that might elicit bNAbs, we produced and immunized mice expressing the predicted germline PGT121, a bNAb specific for the V3-loop and surrounding glycans on the HIV-1 spike. Priming with an epitope-modified immunogen designed to activate germline antibody-expressing B cells, followed by ELISA-guided boosting with a sequence of directional immunogens, native-like trimers with decreasing epitope modification, elicited heterologous tier-2-neutralizing responses. In contrast, repeated immunization with the priming immunogen did not. Antibody cloning confirmed elicitation of high levels of somatic mutation and tier-2-neutralizing antibodies resembling the authentic human bNAb. Our data establish that sequential immunization with specifically designed immunogens can induce high levels of somatic mutation and shepherd antibody maturation to produce bNAbs from their inferred germline precursors.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures



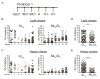
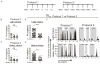


Comment in
-
Teaching a Clone to Walk, One Step at a Time.Cell. 2016 Sep 8;166(6):1360-1361. doi: 10.1016/j.cell.2016.08.039. Cell. 2016. PMID: 27610559
-
HIV Vaccines: One Step Closer.Trends Mol Med. 2017 Jan;23(1):1-3. doi: 10.1016/j.molmed.2016.10.006. Epub 2016 Nov 23. Trends Mol Med. 2017. PMID: 27889424
References
-
- Baumjohann D, Preite S, Reboldi A, Ronchi F, Ansel KM, Lanzavecchia A, Sallusto F. Persistent antigen and germinal center B cells sustain T follicular helper cell responses and phenotype. Immunity. 2013;38:596–605. - PubMed
-
- Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol. 2011;85:9998–10009. - PMC - PubMed
-
- Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5:233–236. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

