Induction of HIV Neutralizing Antibody Lineages in Mice with Diverse Precursor Repertoires
- PMID: 27610571
- PMCID: PMC5103708
- DOI: 10.1016/j.cell.2016.07.029
Induction of HIV Neutralizing Antibody Lineages in Mice with Diverse Precursor Repertoires
Abstract
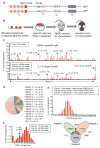
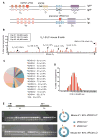
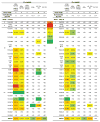
The design of immunogens that elicit broadly reactive neutralizing antibodies (bnAbs) has been a major obstacle to HIV-1 vaccine development. One approach to assess potential immunogens is to use mice expressing precursors of human bnAbs as vaccination models. The bnAbs of the VRC01-class derive from the IGHV1-2 immunoglobulin heavy chain and neutralize a wide spectrum of HIV-1 strains via targeting the CD4 binding site of the envelope glycoprotein gp120. We now describe a mouse vaccination model that allows a germline human IGHV1-2(∗)02 segment to undergo normal V(D)J recombination and, thereby, leads to the generation of peripheral B cells that express a highly diverse repertoire of VRC01-related receptors. When sequentially immunized with modified gp120 glycoproteins designed to engage VRC01 germline and intermediate antibodies, IGHV1-2(∗)02-rearranging mice, which also express a VRC01-antibody precursor light chain, can support the affinity maturation of VRC01 precursor antibodies into HIV-neutralizing antibody lineages.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Teaching a Clone to Walk, One Step at a Time.Cell. 2016 Sep 8;166(6):1360-1361. doi: 10.1016/j.cell.2016.08.039. Cell. 2016. PMID: 27610559
-
HIV Vaccines: One Step Closer.Trends Mol Med. 2017 Jan;23(1):1-3. doi: 10.1016/j.molmed.2016.10.006. Epub 2016 Nov 23. Trends Mol Med. 2017. PMID: 27889424
References
-
- DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, Ellington AD, Georgiou G. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nature medicine. 2015;21:86–91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UM1 AI100645/AI/NIAID NIH HHS/United States
- UM1 AI100663/AI/NIAID NIH HHS/United States
- U19 AI109632/AI/NIAID NIH HHS/United States
- R01 AI020047/AI/NIAID NIH HHS/United States
- F31 AI117920/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 AI077595/AI/NIAID NIH HHS/United States
- R37 AI077595/AI/NIAID NIH HHS/United States
- R01 AI128836/AI/NIAID NIH HHS/United States
- P01 AI104722/AI/NIAID NIH HHS/United States
- R37 AI020047/AI/NIAID NIH HHS/United States
- P01 AI094419/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

