Absolute quantification of microbial taxon abundances
- PMID: 27612291
- PMCID: PMC5270559
- DOI: 10.1038/ismej.2016.117
Absolute quantification of microbial taxon abundances
Abstract
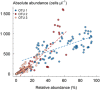
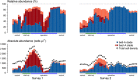
High-throughput amplicon sequencing has become a well-established approach for microbial community profiling. Correlating shifts in the relative abundances of bacterial taxa with environmental gradients is the goal of many microbiome surveys. As the abundances generated by this technology are semi-quantitative by definition, the observed dynamics may not accurately reflect those of the actual taxon densities. We combined the sequencing approach (16S rRNA gene) with robust single-cell enumeration technologies (flow cytometry) to quantify the absolute taxon abundances. A detailed longitudinal analysis of the absolute abundances resulted in distinct abundance profiles that were less ambiguous and expressed in units that can be directly compared across studies. We further provide evidence that the enrichment of taxa (increase in relative abundance) does not necessarily relate to the outgrowth of taxa (increase in absolute abundance). Our results highlight that both relative and absolute abundances should be considered for a comprehensive biological interpretation of microbiome surveys.
Figures
References
-
- Amann R, Fuchs BM. (2008). Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat Rev Microbiol 6: 339–348. - PubMed
-
- Faust K, Lahti L, Gonze D, de Vos WM, Raes J. (2015). Metagenomics meets time series analysis: unraveling microbial community dynamics. Curr Opin Microbiol 25: 56–66. - PubMed
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

