Fast and sequence-adaptive whole-brain segmentation using parametric Bayesian modeling
- PMID: 27612647
- PMCID: PMC8117726
- DOI: 10.1016/j.neuroimage.2016.09.011
Fast and sequence-adaptive whole-brain segmentation using parametric Bayesian modeling
Abstract
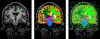
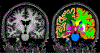
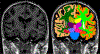
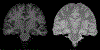
Quantitative analysis of magnetic resonance imaging (MRI) scans of the brain requires accurate automated segmentation of anatomical structures. A desirable feature for such segmentation methods is to be robust against changes in acquisition platform and imaging protocol. In this paper we validate the performance of a segmentation algorithm designed to meet these requirements, building upon generative parametric models previously used in tissue classification. The method is tested on four different datasets acquired with different scanners, field strengths and pulse sequences, demonstrating comparable accuracy to state-of-the-art methods on T1-weighted scans while being one to two orders of magnitude faster. The proposed algorithm is also shown to be robust against small training datasets, and readily handles images with different MRI contrast as well as multi-contrast data.
Keywords: Atlases; Bayesian modeling; MRI; Parametric models; Segmentation.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
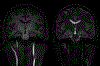
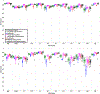
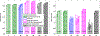
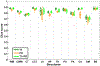
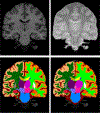
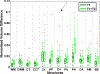
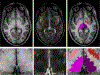
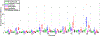
References
-
- Aljabar P, Heckemann AR, Hammers A, Hajnal VJ, Rueckert D, 2009. Multi-atlas based segmentation of brain images: Atlas selection and its effect on accuracy. NeuroImage 46, 726–738. - PubMed
-
- Artaechevarria X, Munõz Barrutia A, Ortiz-de Solórzano C, 2009. Combination strategies in multi-atlas image segmentation: Application to brain MR data. IEEE Transactions on Medical Imaging 28, 1266–1277. - PubMed
-
- Ashburner J, Friston JK, 1997. Multimodal image coregistration and partitioning – a unified framework. NeuroImage 6, 209–217. - PubMed
-
- Ashburner J, Friston JK, 2005. Unified segmentation. NeuroImage 26, 839–885. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

