From single molecules to life: microscopy at the nanoscale
- PMID: 27613013
- PMCID: PMC5566169
- DOI: 10.1007/s00216-016-9781-8
From single molecules to life: microscopy at the nanoscale
Abstract
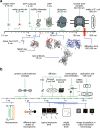
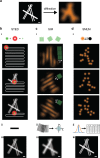
Super-resolution microscopy is the term commonly given to fluorescence microscopy techniques with resolutions that are not limited by the diffraction of light. Since their conception a little over a decade ago, these techniques have quickly become the method of choice for many biologists studying structures and processes of single cells at the nanoscale. In this review, we present the three main approaches used to tackle the diffraction barrier of ∼200 nm: stimulated-emission depletion (STED) microscopy, structured illumination microscopy (SIM), and single-molecule localization microscopy (SMLM). We first present a theoretical overview of the techniques and underlying physics, followed by a practical guide to all of the facets involved in designing a super-resolution experiment, including an approachable explanation of the photochemistry involved, labeling methods available, and sample preparation procedures. Finally, we highlight some of the most exciting recent applications of and developments in these techniques, and discuss the outlook for this field. Graphical Abstract Super-resolution microscopy techniques. Working principles of the common approaches stimulated-emission depletion (STED) microscopy, structured illumination microscopy (SIM), and single-molecule localization microscopy (SMLM).
Keywords: Live cell imaging; Photophysics and photochemistry of fluorophores; Quantitative cell biology; Super-resolution microscopy.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Hell SW, Sahl SJ, Bates M, Zhuang XW, Heintzmann R, Booth MJ et al. The 2015 super-resolution microscopy roadmap. J Phys D Appl Phys. 2015;48(44). doi:10.1088/0022-3727/48/44/443001.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

