Identification of oncogenic driver mutations by genome-wide CRISPR-Cas9 dropout screening
- PMID: 27613601
- PMCID: PMC5016932
- DOI: 10.1186/s12864-016-3042-2
Identification of oncogenic driver mutations by genome-wide CRISPR-Cas9 dropout screening
Abstract
Background: Genome-wide CRISPR-Cas9 dropout screens can identify genes whose knockout affects cell viability. Recent CRISPR screens detected thousands of essential genes required for cellular survival and key cellular processes; however discovering novel lineage-specific genetic dependencies from the many hits still remains a challenge.
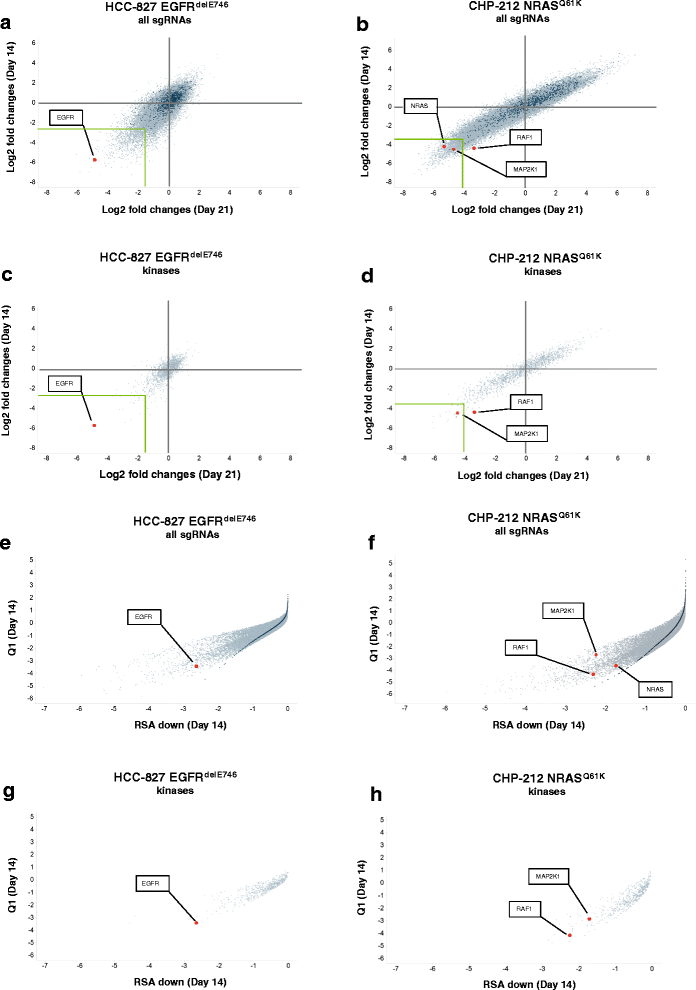
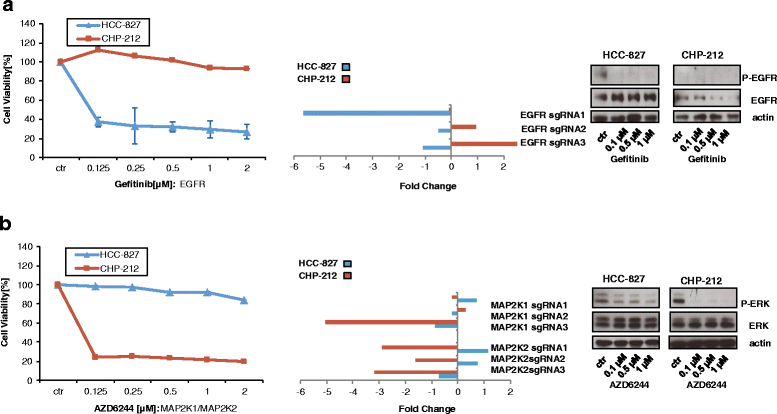
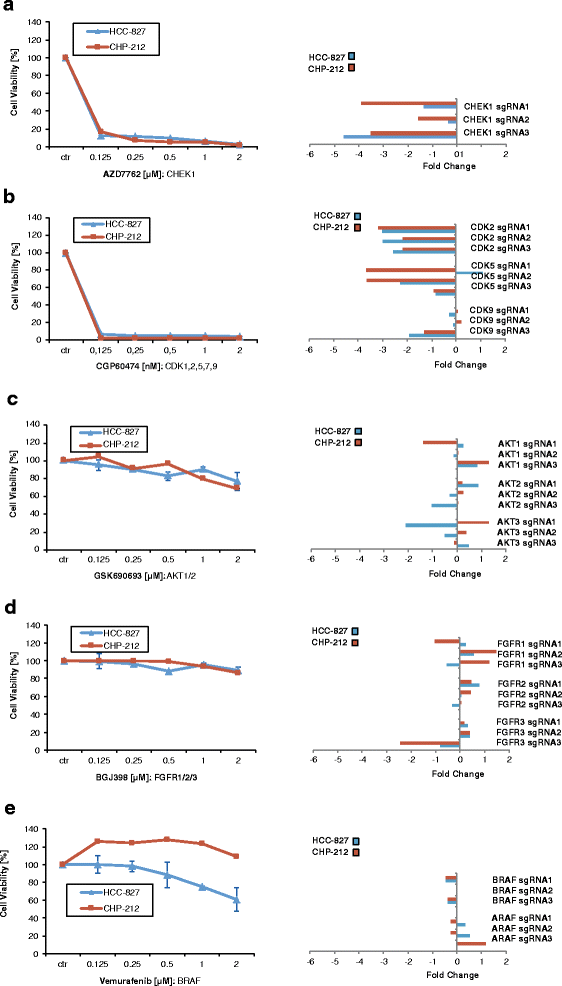
Results: To assess whether CRISPR-Cas9 dropout screens can help identify cancer dependencies, we screened two human cancer cell lines carrying known and distinct oncogenic mutations using a genome-wide sgRNA library. We found that the gRNA targeting the driver mutation EGFR was one of the highest-ranking candidates in the EGFR-mutant HCC-827 lung adenocarcinoma cell line. Likewise, sgRNAs for NRAS and MAP2K1 (MEK1), a downstream kinase of mutant NRAS, were identified among the top hits in the NRAS-mutant neuroblastoma cell line CHP-212. Depletion of these genes targeted by the sgRNAs strongly correlated with the sensitivity to specific kinase inhibitors of the EGFR or RAS pathway in cell viability assays. In addition, we describe other dependencies such as TBK1 in HCC-827 cells and TRIB2 in CHP-212 cells which merit further investigation.
Conclusions: We show that genome-wide CRISPR dropout screens are suitable for the identification of oncogenic drivers and other essential genes.
Keywords: Driver mutations; Dropout; EGFR; Kinase; NRAS; Negative selection; Whole genome CRISPR screen.
Figures


References
-
- Hart T, et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell. 2015;163(6):1515-26. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

