Effect of X-ray irradiation on ancient DNA in sub-fossil bones - Guidelines for safe X-ray imaging
- PMID: 27615365
- PMCID: PMC5018823
- DOI: 10.1038/srep32969
Effect of X-ray irradiation on ancient DNA in sub-fossil bones - Guidelines for safe X-ray imaging
Abstract
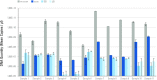
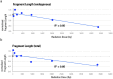
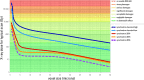
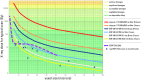
Sub-fossilised remains may still contain highly degraded ancient DNA (aDNA) useful for palaeogenetic investigations. Whether X-ray computed [micro-] tomography ([μ]CT) imaging of these fossils may further damage aDNA remains debated. Although the effect of X-ray on DNA in living organisms is well documented, its impact on aDNA molecules is unexplored. Here we investigate the effects of synchrotron X-ray irradiation on aDNA from Pleistocene bones. A clear correlation appears between decreasing aDNA quantities and accumulating X-ray dose-levels above 2000 Gray (Gy). We further find that strong X-ray irradiation reduces the amount of nucleotide misincorporations at the aDNA molecule ends. No representative effect can be detected for doses below 200 Gy. Dosimetry shows that conventional μCT usually does not reach the risky dose level, while classical synchrotron imaging can degrade aDNA significantly. Optimised synchrotron protocols and simple rules introduced here are sufficient to ensure that fossils can be scanned without impairing future aDNA studies.
Figures




References
-
- Hublin J. J. et al.. Suggested guidelines for invasive sampling of hominid remains. J Hum Evol 55, 756–757 (2008). - PubMed
-
- Richards G. D., Jabbour R. S., Horton C. F., Ibarra C. L. & MacDowell A. A. Color changes in modern and fossil teeth induced by synchrotron microtomography. Am J Phys Anthropol 149, 172–180 (2012). - PubMed
-
- Tafforeau P. & Smith T. M. Nondestructive imaging of hominoid dental microstructure using phase contrast X-ray synchrotron microtomography. J Hum Evol 54, 272–278 (2008). - PubMed
-
- Teoule R. Radiation-induced DNA damage and its repair. Int J Radiat Biol Relat Stud Phys Chem Med 51, 573–589 (1987). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

