Interactions between host genetics and gut microbiome in diabetes and metabolic syndrome
- PMID: 27617202
- PMCID: PMC5004229
- DOI: 10.1016/j.molmet.2016.07.004
Interactions between host genetics and gut microbiome in diabetes and metabolic syndrome
Abstract
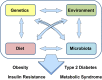
Background: Diabetes, obesity, and the metabolic syndrome are multifactorial diseases dependent on a complex interaction of host genetics, diet, and other environmental factors. Increasing evidence places gut microbiota as important modulators of the crosstalk between diet and development of obesity and metabolic dysfunction. In addition, host genetics can have important impact on the composition and function of gut microbiota. Indeed, depending on the genetic background of the host, diet and other environmental factors may produce different changes in gut microbiota, have different impacts on host metabolism, and create different interactions between the microbiome and the host.
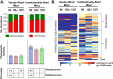
Scope of review: In this review, we highlight how appropriate animal models can help dissect the complex interaction of host genetics with the gut microbiome and how diet can lead to different degrees of weight gain, levels of insulin resistance, and metabolic outcomes, such as diabetes, in different individuals. We also discuss the challenges of identifying specific disease-associated microbiota and the limitations of simple metrics, such as phylogenetic diversity or the ratio of Firmicutes to Bacteroidetes.
Major conclusions: Understanding these complex interactions will help in the development of novel treatments for microbiome-related metabolic diseases. This article is part of a special issue on microbiota.
Keywords: Environment; Host genetics; Metabolic syndrome; Microbial diversity; Microbiome; Microbiota; Obesity.
Figures

References
-
- Tremaroli V., Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

