Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy
- PMID: 27617430
- PMCID: PMC5515730
- DOI: 10.1038/nsmb.3293
Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy
Abstract
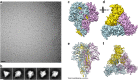
The threat of a major coronavirus pandemic urges the development of strategies to combat these pathogens. Human coronavirus NL63 (HCoV-NL63) is an α-coronavirus that can cause severe lower-respiratory-tract infections requiring hospitalization. We report here the 3.4-Å-resolution cryo-EM reconstruction of the HCoV-NL63 coronavirus spike glycoprotein trimer, which mediates entry into host cells and is the main target of neutralizing antibodies during infection. The map resolves the extensive glycan shield obstructing the protein surface and, in combination with mass spectrometry, provides a structural framework to understand the accessibility to antibodies. The structure reveals the complete architecture of the fusion machinery including the triggering loop and the C-terminal domains, which contribute to anchoring the trimer to the viral membrane. Our data further suggest that HCoV-NL63 and other coronaviruses use molecular trickery, based on epitope masking with glycans and activating conformational changes, to evade the immune system of infected hosts.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



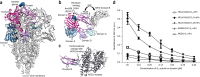
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

